
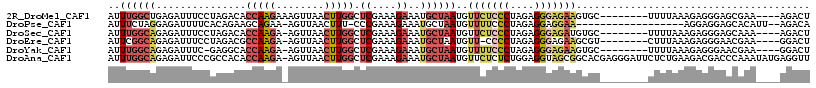
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,582,155 – 8,582,260 |
| Length | 105 |
| Max. P | 0.920615 |

| Location | 8,582,155 – 8,582,260 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.75 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -12.44 |
| Energy contribution | -14.17 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8582155 105 + 20766785 AUUUGGCUGAGAUUUCCUAGACACCAAGAAAGUUAACUUGGCUCGAAAGAAAUGCUAAUGUUCUCCCUAGAGGGAGAAGUGC--------UUUUAAAGAGGGAGCGAA----AGACU ..(((((................(((((........))))).((....))...)))))..(((((((....))))))).(((--------((((.....)))))))..----..... ( -27.10) >DroPse_CAF1 15927 95 + 1 AUUUCUAGGAGAUUUUCACAGAAGCAGAA-AGUUAACUUU-CCCGAAAGAAAUGCUAAUGUUUUCCCUAGAGGAGGAA------------------AGGAGGAGCACAUU--AGACA .((((((((.((.....(((..(((((((-((....))))-).(....)...))))..)))..)))))))))).....------------------..............--..... ( -21.40) >DroSec_CAF1 57624 104 + 1 AUUUGGCAGAGAUUUCCUAGACACCAAGA-AGUUAACUUGGCUCGAAAGAAAUGCUAAUGUUCUCCCUAGAGGGAGAUGUGC--------UUUUAAAGAGGGAGCAAA----AGACU ..((((((...............(((((.-......))))).((....))..))))))...((((((....))))))..(((--------((((.....)))))))..----..... ( -27.90) >DroEre_CAF1 48444 103 + 1 AUUCGGCAGAGAUUUCCUAGACGCCAAGA-AGUUAACUUGGCUCGAAAGAAAUGCUAAUGUU-CCCCUAGAGGGAGAAGCGU--------CUUUAAAGAGGGAACGAA----GGACU .(((((((....((((...((.((((((.-......))))))))))))....))))...(((-(((((..(((((......)--------))))..)).)))))))))----..... ( -30.70) >DroYak_CAF1 61166 103 + 1 AUUUGGCAGAGAUUUC-GAGGCACCAAGA-AGUUAACUUGGCUCGAAAGAAAUGCUAAUGUUUUCCCUAGAGGGAGAAGUGC--------UUUUAAAGAGGGAACGAA----GGACU ..((((((....((((-(((...(((((.-......))))))))))))....))))))..(((((((....)))))))((.(--------(((............)))----).)). ( -30.50) >DroAna_CAF1 8368 116 + 1 AUUUGGCAGAGAUUCCCGCCACACCAAGA-AGUUAACUUGGCUCGAAAGAAAUGCUAAUGUUCUCUCUGGAGGUAGCGGCACGAGGGAUUCUCUGAAGACGACCCAAAUAUGAGGUU ..((..((((((.(((((((.((((.(((-.(..((((((((((....))...))))).)))..))))...))).).)))....)))).))))))..)).((((((....)).)))) ( -37.70) >consensus AUUUGGCAGAGAUUUCCUAGACACCAAGA_AGUUAACUUGGCUCGAAAGAAAUGCUAAUGUUCUCCCUAGAGGGAGAAGUGC________UUUUAAAGAGGGAGCAAA____AGACU ..((((((...............(((((........))))).((....))..))))))..(((((((....)))))))....................................... (-12.44 = -14.17 + 1.73)


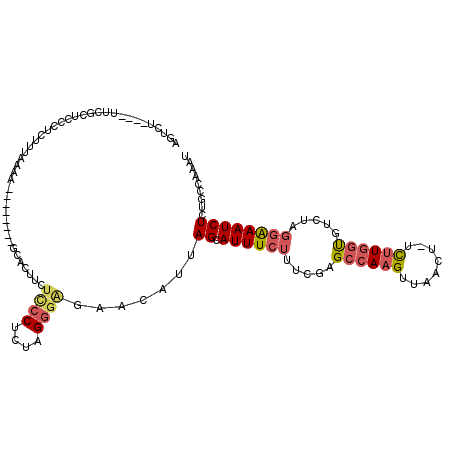
| Location | 8,582,155 – 8,582,260 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.75 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -11.17 |
| Energy contribution | -11.87 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8582155 105 - 20766785 AGUCU----UUCGCUCCCUCUUUAAAA--------GCACUUCUCCCUCUAGGGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACUUUCUUGGUGUCUAGGAAAUCUCAGCCAAAU .....----...(((............--------((..(((((((....))))))).....))(((((((....((((((........))))))....)))))))...)))..... ( -26.10) >DroPse_CAF1 15927 95 - 1 UGUCU--AAUGUGCUCCUCCU------------------UUCCUCCUCUAGGGAAAACAUUAGCAUUUCUUUCGGG-AAAGUUAACU-UUCUGCUUCUGUGAAAAUCUCCUAGAAAU ((.((--(((((..((((...------------------...........))))..))))))))).........((-((((....))-))))..(((((.((.....)).))))).. ( -19.34) >DroSec_CAF1 57624 104 - 1 AGUCU----UUUGCUCCCUCUUUAAAA--------GCACAUCUCCCUCUAGGGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACU-UCUUGGUGUCUAGGAAAUCUCUGCCAAAU .((((----((((.........)))))--------).)).((((((....))))))......(((((((((....((((((......-.))))))....))))))....)))..... ( -25.90) >DroEre_CAF1 48444 103 - 1 AGUCC----UUCGUUCCCUCUUUAAAG--------ACGCUUCUCCCUCUAGGGG-AACAUUAGCAUUUCUUUCGAGCCAAGUUAACU-UCUUGGCGUCUAGGAAAUCUCUGCCGAAU .....----((((((((((......((--------(....))).......))))-)))....(((((((((....((((((......-.))))))....))))))....))).))). ( -26.52) >DroYak_CAF1 61166 103 - 1 AGUCC----UUCGUUCCCUCUUUAAAA--------GCACUUCUCCCUCUAGGGAAAACAUUAGCAUUUCUUUCGAGCCAAGUUAACU-UCUUGGUGCCUC-GAAAUCUCUGCCAAAU .....----..................--------.......((((....))))........(((....((((((((((((......-.)))))...)))-))))....)))..... ( -20.20) >DroAna_CAF1 8368 116 - 1 AACCUCAUAUUUGGGUCGUCUUCAGAGAAUCCCUCGUGCCGCUACCUCCAGAGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACU-UCUUGGUGUGGCGGGAAUCUCUGCCAAAU ........((((((........((((((.((((....(((((........(((((((.........)))))))..((((((......-.))))))))))))))).)))))))))))) ( -37.70) >consensus AGUCU____UUCGCUCCCUCUUUAAAA________GCACUUCUCCCUCUAGGGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACU_UCUUGGUGUCUAGGAAAUCUCUGCCAAAU ..........................................((((....)))).......((.((((((.....((((((........)))))).....))))))))......... (-11.17 = -11.87 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:02 2006