
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,569,091 – 8,569,225 |
| Length | 134 |
| Max. P | 0.998500 |

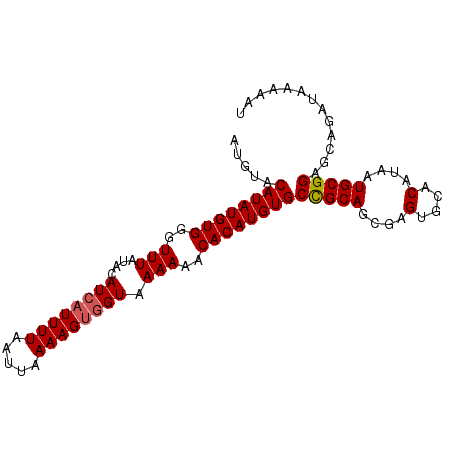
| Location | 8,569,091 – 8,569,185 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -19.34 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8569091 94 + 20766785 AUGUACAUAUGUGGAUUUAUACAUAAUUUUAAUUAAAAGUGGUAAAAAACACAUGUGCCGCAGCGAGUGCACAUAAUGCGGUGCAGAUAAAAAU .((((((((((((..(((.(((...(((((.....))))).))))))..))))))))(((((....(....)....)))))))))......... ( -20.90) >DroSec_CAF1 44737 94 + 1 AUGUACAUAUGUGGGUUUAUACAUCAUUUUAAUUAAAAGUGGUAAAAAACACAUGUGCCGCAGCGAGUGCACAUAAUGCGGAGCAGAUAAAAAU .(((.((((((((..(((....((((((((.....)))))))).)))..))))))))(((((....(....)....))))).)))......... ( -22.80) >DroSim_CAF1 48421 94 + 1 AUGUACAUAUGUGGGUUUAUACAUCAUUUUAAUUAAAAGUGGUAAAAAACACAUGUGCCGCAGCGAGUGCACAUAAUGCGGAGCAGAUAAAAAU .(((.((((((((..(((....((((((((.....)))))))).)))..))))))))(((((....(....)....))))).)))......... ( -22.80) >DroEre_CAF1 35773 94 + 1 AUGUACAUAUGUGGGUUUGUACAUCAUUUUAAUUAAAAGUGGUAAAAAACACAUGUGCUGCAGCGAGUGCACAUAAUGCGGAGCAGAUAAAAAU .((((((((((((..(((....((((((((.....)))))))).)))..)))))))).))))((...((((.....))))..)).......... ( -24.40) >DroYak_CAF1 48344 94 + 1 AUAUACAUAUGUGGGUUUGUACAUCAUUUUAAUUAAAAGCGGUAAAAAACACAUGUGCUGCAGGCAGUGCACAUAAUGCGGAGCAGAUAAAACU .....((((((((..(((.(((....((((....))))...))))))..))))))))((((..(((..........)))...))))........ ( -19.50) >consensus AUGUACAUAUGUGGGUUUAUACAUCAUUUUAAUUAAAAGUGGUAAAAAACACAUGUGCCGCAGCGAGUGCACAUAAUGCGGAGCAGAUAAAAAU .....((((((((..(((....((((((((.....)))))))).)))..))))))))(((((....(....)....)))))............. (-19.34 = -19.50 + 0.16)


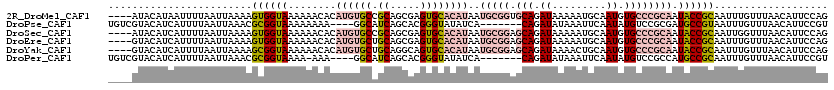
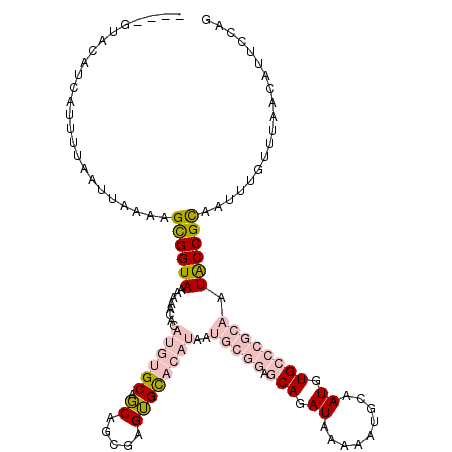
| Location | 8,569,109 – 8,569,225 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -16.66 |
| Energy contribution | -18.72 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8569109 116 + 20766785 ----AUACAUAAUUUUAAUUAAAAGUGGUAAAAAACACAUGUGCCGCAGCGAGUGCACAUAAUGCGGUGCAGAUAAAAAUGCAAUGUGCCCGCAAUACCGCAAUUUGUUUAACAUUCCAG ----....................((((((........((((((.((.....))))))))..(((((.(((.((.........)).)))))))).))))))................... ( -28.40) >DroPse_CAF1 2778 109 + 1 UGUCGUACAUCAUUUUAAUUAAACGCGGUAAAAAAAA----GGCAUCAGCACGGGUAUAUCA-------CAGAUAUAAAUUCAAUAUGUCCGCGAUGCCGUAAUUUGUUUAACAUUCCGU ........................((((.........----((((((.((..((..((((..-------..((.......)).))))..))))))))))......((.....))..)))) ( -18.80) >DroSec_CAF1 44755 116 + 1 ----AUACAUCAUUUUAAUUAAAAGUGGUAAAAAACACAUGUGCCGCAGCGAGUGCACAUAAUGCGGAGCAGAUAAAAAUGCAAUGUGCCCGCAAUACCGCAAUUGGUUUAACAUUCCAG ----....................((((((........((((((.((.....))))))))..(((((.(((.((.........)).)))))))).))))))...(((.........))). ( -29.40) >DroEre_CAF1 35791 116 + 1 ----GUACAUCAUUUUAAUUAAAAGUGGUAAAAAACACAUGUGCUGCAGCGAGUGCACAUAAUGCGGAGCAGAUAAAAAUGCAAUGUGCCCGCAAUACCGCAAUUUGUUUAACAUUCCAG ----....................((((((........((((((.((.....))))))))..(((((.(((.((.........)).)))))))).))))))................... ( -28.40) >DroYak_CAF1 48362 116 + 1 ----GUACAUCAUUUUAAUUAAAAGCGGUAAAAAACACAUGUGCUGCAGGCAGUGCACAUAAUGCGGAGCAGAUAAAACUGCAAUGUGCCCGCAAUACCGCAAUUUGUUUAACAUUCCAG ----....................((((((.........((..(((....)))..)).....(((((.(((.((.........)).)))))))).))))))................... ( -31.20) >DroPer_CAF1 3032 108 + 1 UGUCGUACAUCAUUUUAAUUAAACGCGGUAAAA-AAA----GGCAUCAGCACGGGUAUAUCA-------CAGAUAUAAAUUCAAUAUGUCCGCCAUGCCGCAAUUUGUUUAACAUUCCGU ..................((((((((((((...-...----.((....)).(((..((((..-------..((.......)).))))..)))...)))))).....))))))........ ( -18.00) >consensus ____GUACAUCAUUUUAAUUAAAAGCGGUAAAAAACACAUGUGCAGCAGCGAGUGCACAUAAUGCGGAGCAGAUAAAAAUGCAAUGUGCCCGCAAUACCGCAAUUUGUUUAACAUUCCAG ........................((((((........((((((.((.....))))))))..(((((.(((.((.........)).)))))))).))))))................... (-16.66 = -18.72 + 2.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:58 2006