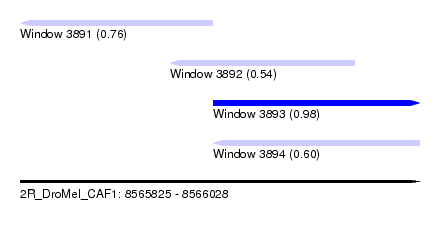
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,565,825 – 8,566,028 |
| Length | 203 |
| Max. P | 0.982646 |

| Location | 8,565,825 – 8,565,923 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 94.52 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -22.34 |
| Energy contribution | -22.57 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8565825 98 - 20766785 UUAUCUAACAUCCUGUUUUUAAGGAAUCCAAAAUGGUCUUAAAGUGGGCUUAGAGUAACGAAAGUGUUUCGACUGUCAGAAUGACGAAAGCAUAAAAG ...(((.((.((((.......)))).........((((...(((....))).(((..((....))..))))))))).)))(((.(....))))..... ( -17.90) >DroSec_CAF1 41500 96 - 1 CUAUCUAACAUCCUGUUUUUAAGGAAUCCA-AAUGGUCUUA-AGUGGGCUCAGAGAUACGAAAGUGUUUCGACUGUCAGAAUGACGAAAGCAUAAAAG ...(((.(((((((.......)))).....-..((((((..-...)))).))(((((((....)))))))...))).)))(((.(....))))..... ( -23.60) >DroSim_CAF1 44773 96 - 1 CUAUCUAGCAUCCUGUUUUUAAGGAAUCCA-AAUGGUCUUA-AGUGGACUUAGAGAUACGAAAGUGUUUCGACUGUCAGAAUGACGAAAGCAUAAAAG ...(((..(.((((.......)))).....-...((((.((-((....))))(((((((....))))))))))))..)))(((.(....))))..... ( -25.50) >consensus CUAUCUAACAUCCUGUUUUUAAGGAAUCCA_AAUGGUCUUA_AGUGGGCUUAGAGAUACGAAAGUGUUUCGACUGUCAGAAUGACGAAAGCAUAAAAG ...(((.(((((((.......)))).........(((((......)))))..(((((((....)))))))...))).)))(((.(....))))..... (-22.34 = -22.57 + 0.23)

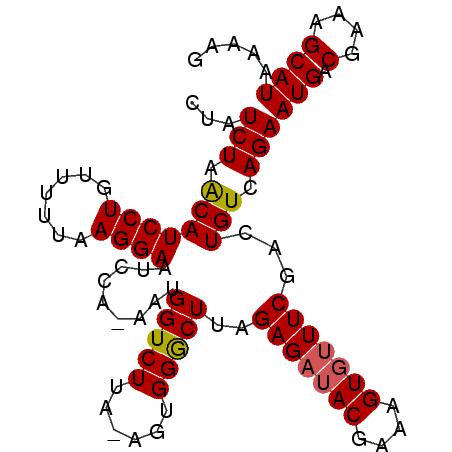
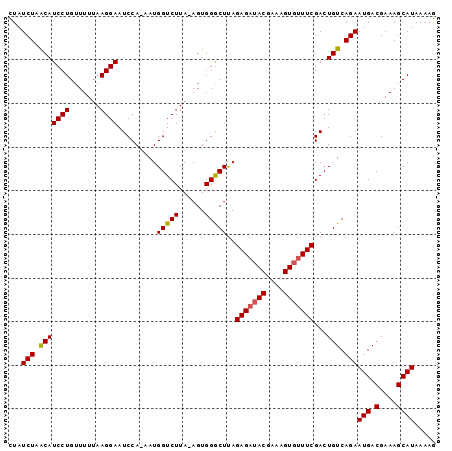
| Location | 8,565,901 – 8,565,995 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.34 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -16.22 |
| Energy contribution | -16.26 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8565901 94 - 20766785 GCCAAUUGUCGAAUCUGACGAUGGUAAGUGCAUGUGUGUGCACUGAAAACAAAUAGUGUACAAUCGCCUUUA--------------------UUAUCUAACAUCCUGUUUUUAA .......((((....))))(((((((((.((.....(((((((((........)))))))))...)).))))--------------------)))))................. ( -22.10) >DroSec_CAF1 41574 94 - 1 GCCAAUUGUCGAAUCUGGCGAUGGUAAGUGCAUGUGUGUGCACUGAAAACAAAUGGUGUACAAAGCUCUUCA--------------------CUAUCUAACAUCCUGUUUUUAA ((((..(.....)..))))(((((((((.((.....((((((((..........))))))))..)).))).)--------------------)))))................. ( -24.40) >DroSim_CAF1 44847 94 - 1 GCCAAUUGUCGAAUCUGGCGAUGGUAAGUGCAUGUGUGUGCACUGAAAACAAAUAGUGUACAAAGCUCUUUA--------------------CUAUCUAGCAUCCUGUUUUUAA ((((..(.....)..))))(((((((((.((.....(((((((((........)))))))))..)).).)))--------------------)))))................. ( -25.40) >DroEre_CAF1 32634 100 - 1 GCCAAUUGUCGAAUCUGGCGAUGCUAAGUGCAUGUGUGUGCACUGAAAACAAAUUGCUGAGGCAACUCAUUAUA--------UACUGUAAAA------GGCGUCCUGUUUGUGU ((((..(.....)..))))((((((...((((.(((((((((.(........).)))((((....))))..)))--------)))))))...------)))))).......... ( -27.90) >DroYak_CAF1 45075 114 - 1 GCCAAUUGUCGAAUCUGGCGAUGGUAAGUGCAUGUGUGUGCACUGAAAACAAAUUGUUAAGCCAACUCAUUAUAACGCCUUAUACCCCUAGAUUGCUCAGCAUCCUGAAUUAAU ((((.((((((....)))))))))).(((((((....))))))).....((...((((.(((...((...(((((....))))).....))...))).))))...))....... ( -21.90) >consensus GCCAAUUGUCGAAUCUGGCGAUGGUAAGUGCAUGUGUGUGCACUGAAAACAAAUAGUGUACAAAACUCUUUA____________________CUAUCUAGCAUCCUGUUUUUAA ((((.((((((....)))))))))).(((((((....)))))))...................................................................... (-16.22 = -16.26 + 0.04)

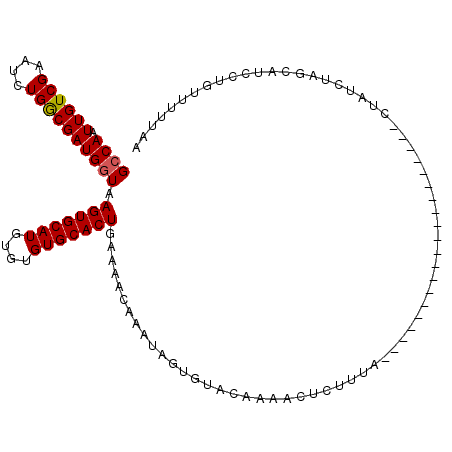
| Location | 8,565,923 – 8,566,028 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8565923 105 + 20766785 --UAAAGGCGAUUGUACACUAUUUGUUUUCAGUGCACACACAUGCACUUACCAUCGUCAGAUUCGACAAUUGGCAAAUUUAAAAGCAAUAAACGUACAGAGAGCAAA --.....((..((((((.....(((((((.((((((......))))))..(((..(((......)))...)))........))))))).....))))))...))... ( -23.50) >DroSec_CAF1 41596 105 + 1 --UGAAGAGCUUUGUACACCAUUUGUUUUCAGUGCACACACAUGCACUUACCAUCGCCAGAUUCGACAAUUGGCAAAUUUAAAAGCAAUAAACGUACAGAGAGCAAA --....(..((((((((.....(((((((.((((((......)))))).......(((((.........))))).......))))))).....))))))))..)... ( -27.00) >DroSim_CAF1 44869 105 + 1 --UAAAGAGCUUUGUACACUAUUUGUUUUCAGUGCACACACAUGCACUUACCAUCGCCAGAUUCGACAAUUGGCAAAUUUAAAAGCAAUAAACGUACAGAGAGCAAA --....(..((((((((.....(((((((.((((((......)))))).......(((((.........))))).......))))))).....))))))))..)... ( -27.00) >DroEre_CAF1 32660 107 + 1 UAUAAUGAGUUGCCUCAGCAAUUUGUUUUCAGUGCACACACAUGCACUUAGCAUCGCCAGAUUCGACAAUUGGCAAAUUUAAAAGCAAUAAACGUACAGAGAGCAAA ......(((((((....)))))))((((((.((((......((((.....)))).(((((.........)))))...................)))).))))))... ( -23.80) >DroYak_CAF1 45115 107 + 1 UAUAAUGAGUUGGCUUAACAAUUUGUUUUCAGUGCACACACAUGCACUUACCAUCGCCAGAUUCGACAAUUGGCAAAUUUAAAAGCAAUAAACGUACAGAGAGCAAA ......((((((......))))))((((((((((((......)))))).......(((((.........)))))........................))))))... ( -21.40) >consensus __UAAAGAGCUUUGUACACAAUUUGUUUUCAGUGCACACACAUGCACUUACCAUCGCCAGAUUCGACAAUUGGCAAAUUUAAAAGCAAUAAACGUACAGAGAGCAAA .....................(((((((((((((((......)))))).......(((((.........)))))........................))))))))) (-19.32 = -19.16 + -0.16)


| Location | 8,565,923 – 8,566,028 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -20.12 |
| Energy contribution | -20.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8565923 105 - 20766785 UUUGCUCUCUGUACGUUUAUUGCUUUUAAAUUUGCCAAUUGUCGAAUCUGACGAUGGUAAGUGCAUGUGUGUGCACUGAAAACAAAUAGUGUACAAUCGCCUUUA-- ...((....((((((((..(((.(((((...((((((.((((((....))))))))))))((((((....))))))))))).)))..))))))))...)).....-- ( -27.10) >DroSec_CAF1 41596 105 - 1 UUUGCUCUCUGUACGUUUAUUGCUUUUAAAUUUGCCAAUUGUCGAAUCUGGCGAUGGUAAGUGCAUGUGUGUGCACUGAAAACAAAUGGUGUACAAAGCUCUUCA-- ...(((...((((((((..(((.(((((...((((((.((((((....))))))))))))((((((....))))))))))).)))..)))))))).)))......-- ( -28.20) >DroSim_CAF1 44869 105 - 1 UUUGCUCUCUGUACGUUUAUUGCUUUUAAAUUUGCCAAUUGUCGAAUCUGGCGAUGGUAAGUGCAUGUGUGUGCACUGAAAACAAAUAGUGUACAAAGCUCUUUA-- ...(((...((((((((..(((.(((((...((((((.((((((....))))))))))))((((((....))))))))))).)))..)))))))).)))......-- ( -28.30) >DroEre_CAF1 32660 107 - 1 UUUGCUCUCUGUACGUUUAUUGCUUUUAAAUUUGCCAAUUGUCGAAUCUGGCGAUGCUAAGUGCAUGUGUGUGCACUGAAAACAAAUUGCUGAGGCAACUCAUUAUA ((((.((..((((((..(((.((.((((...((((((..(.....)..))))))...)))).)))))..))))))..))...))))....((((....))))..... ( -24.90) >DroYak_CAF1 45115 107 - 1 UUUGCUCUCUGUACGUUUAUUGCUUUUAAAUUUGCCAAUUGUCGAAUCUGGCGAUGGUAAGUGCAUGUGUGUGCACUGAAAACAAAUUGUUAAGCCAACUCAUUAUA ..(((.....)))........((((...(((((((((.((((((....)))))))))..(((((((....))))))).....))))))...))))............ ( -23.30) >consensus UUUGCUCUCUGUACGUUUAUUGCUUUUAAAUUUGCCAAUUGUCGAAUCUGGCGAUGGUAAGUGCAUGUGUGUGCACUGAAAACAAAUAGUGUACAAAACUCUUUA__ ((((.((..((((((..(((.((......((((((((.((((((....)))))))))))))))))))..))))))..))...))))..................... (-20.12 = -20.16 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:55 2006