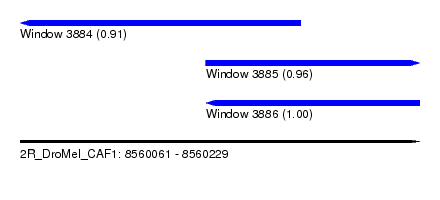
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,560,061 – 8,560,229 |
| Length | 168 |
| Max. P | 0.995106 |

| Location | 8,560,061 – 8,560,179 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 98.31 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.94 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
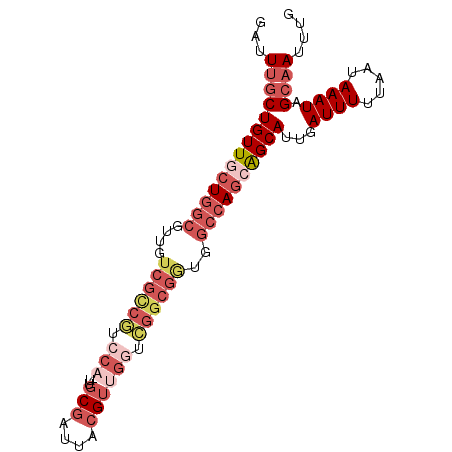
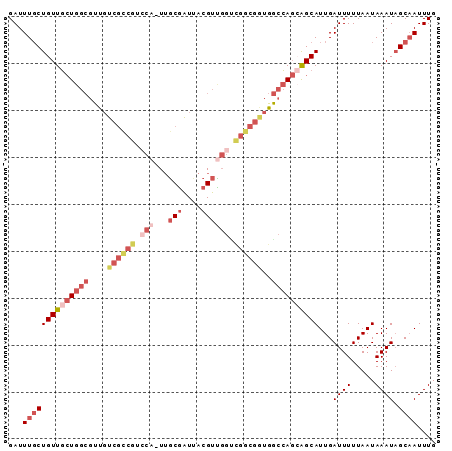
>2R_DroMel_CAF1 8560061 118 - 20766785 CGGUGGCCAGCAGCAUUGAUUUUUAAUAAAUAGCAAUUUGGCAAACCAAUCUGAGCUGUGAUUAUUUUAGCUGGCAGGUGGAAAAAGCAUUUCAAAUGUGCAGCGGACAAGGCCAAAA ...(((((.(((((....((((.....))))..((..((((....))))..)).)))))..........(((((((..(((((......)))))..))).))))......)))))... ( -32.40) >DroSec_CAF1 35803 118 - 1 CGAUGGCCAGCAGCAUUGAUUUUUAAUAAAUAGCAAUUUGGCAAACCAAUCUGAGCUGUGAUUAUUUUAGCUGGCAGGUGGAAAAAGCAUUUCAAAUGUGCAGCGGACAAGGCCAAAA ...(((((.(((((....((((.....))))..((..((((....))))..)).)))))..........(((((((..(((((......)))))..))).))))......)))))... ( -32.50) >DroSim_CAF1 39096 118 - 1 CGGUGGCCAGAAGCAUUGAUUUUUAAUAAAUAGCAAUUUGGCAAACCAAUCUGAGCUGUGAUUAUUUUAGCUGGCAGGUGGAAAAAGCAUUUCAAAUGUGCAGCGGACAAGGCCAAAA ...(((((.....................(((((...((((....)))).....)))))..........(((((((..(((((......)))))..))).))))......)))))... ( -29.00) >DroEre_CAF1 24539 118 - 1 CGGUGGCCAGCAGCAUUGAUUUUUAAUAAAUAGCAAUUUGGCAAACCAAUCUGAGUUGUGAUUAUUUUAGCUGGCAGGUGGAAAAAGCAUUUCAAAUGUGCAGCGGACAAGGCGAAAA .....(((.(((((....((((.....))))..((..((((....))))..)).)))))..........(((((((..(((((......)))))..))).))))......)))..... ( -25.80) >DroYak_CAF1 39083 118 - 1 CGGUGGCCAGCAGCAUUGAUUUUUAAUAAAUAGCAAUUUGGCAAACCAAUCUGGGCUGUGAUUAUUUUAGCUGGCAGGUGGAAAAAGCAUUUCAAAUGUGCAGCGGACAAGGCCAAAA ...(((((.(((((....((((.....))))..((..((((....))))..)).)))))..........(((((((..(((((......)))))..))).))))......)))))... ( -32.40) >consensus CGGUGGCCAGCAGCAUUGAUUUUUAAUAAAUAGCAAUUUGGCAAACCAAUCUGAGCUGUGAUUAUUUUAGCUGGCAGGUGGAAAAAGCAUUUCAAAUGUGCAGCGGACAAGGCCAAAA ...(((((.(((((....((((.....))))..((..((((....))))..)).)))))..........(((((((..(((((......)))))..))).))))......)))))... (-29.70 = -29.94 + 0.24)

| Location | 8,560,139 – 8,560,229 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -21.76 |
| Consensus MFE | -10.88 |
| Energy contribution | -14.00 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8560139 90 + 20766785 CAAAUUGCUAUUUAUUAAAAAUCAAUGCUGCUGGCCACCGCCGGCCAACGUAAUCGCAA-UGGACGGCGACAACGCCAGCAACAGCAAAUC ....(((((((((.....))))....(.(((((((...(((((.(((.((....))...-))).))))).....))))))).))))))... ( -27.70) >DroSec_CAF1 35881 90 + 1 CAAAUUGCUAUUUAUUAAAAAUCAAUGCUGCUGGCCAUCGCCGACCAACGUAAUCGCAA-UGGACGGCGACAACGCCAGCAACAGCAAAUC ....(((((((((.....))))....(.(((((((..((((((.(((.((....))...-))).))))))....))))))).))))))... ( -29.30) >DroSim_CAF1 39174 90 + 1 CAAAUUGCUAUUUAUUAAAAAUCAAUGCUUCUGGCCACCGCCGACCAACGUAAUCGCAA-UGGACGGCGACAACGCCAGCAACAGCAAAUC .........................((((.(((((...(((((.(((.((....))...-))).))))).....)))))....)))).... ( -22.80) >DroEre_CAF1 24617 88 + 1 CAAAUUGCUAUUUAUUAAAAAUCAAUGCUGCUGGCCACCGA---GCGACGUGAUCGCAAAAGGACGACGACAACGCCAGCAACAGCAAAUC ....(((((((((.....))))....(.(((((((......---((((.....))))....(.....)......))))))).))))))... ( -19.40) >DroWil_CAF1 3973 79 + 1 CAAAUGGCAAUUUAUUAAAAAUCAAUGCCUGUCGUU----CCAACACACAAAACAGG-A-UG--UGAAGAGAA---UA-GAACAGAAAAUC ..........((((((.....(((...(((((.((.----.......))...)))))-.-..--)))....))---))-)).......... ( -9.60) >consensus CAAAUUGCUAUUUAUUAAAAAUCAAUGCUGCUGGCCACCGCCGACCAACGUAAUCGCAA_UGGACGGCGACAACGCCAGCAACAGCAAAUC .........................((.(((((((...(((((.(((.((....))....))).))))).....))))))).))....... (-10.88 = -14.00 + 3.12)


| Location | 8,560,139 – 8,560,229 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -17.92 |
| Energy contribution | -21.48 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8560139 90 - 20766785 GAUUUGCUGUUGCUGGCGUUGUCGCCGUCCA-UUGCGAUUACGUUGGCCGGCGGUGGCCAGCAGCAUUGAUUUUUAAUAAAUAGCAAUUUG ...((((((((((((((..(..(((((.(((-..(((....)))))).)))))..))))))))))....((((.....))))))))).... ( -36.50) >DroSec_CAF1 35881 90 - 1 GAUUUGCUGUUGCUGGCGUUGUCGCCGUCCA-UUGCGAUUACGUUGGUCGGCGAUGGCCAGCAGCAUUGAUUUUUAAUAAAUAGCAAUUUG ...((((((((((((((...(((((((.(((-..(((....)))))).))))))).)))))))))....((((.....))))))))).... ( -39.10) >DroSim_CAF1 39174 90 - 1 GAUUUGCUGUUGCUGGCGUUGUCGCCGUCCA-UUGCGAUUACGUUGGUCGGCGGUGGCCAGAAGCAUUGAUUUUUAAUAAAUAGCAAUUUG ...(((((((((((.(.((..((((((.(((-..(((....)))))).))))))..)))...)))(((((...))))).)))))))).... ( -32.80) >DroEre_CAF1 24617 88 - 1 GAUUUGCUGUUGCUGGCGUUGUCGUCGUCCUUUUGCGAUCACGUCGC---UCGGUGGCCAGCAGCAUUGAUUUUUAAUAAAUAGCAAUUUG ...((((((((((((((...((.(((((......))))).))..(((---...))))))))))))....((((.....))))))))).... ( -26.70) >DroWil_CAF1 3973 79 - 1 GAUUUUCUGUUC-UA---UUCUCUUCA--CA-U-CCUGUUUUGUGUGUUGG----AACGACAGGCAUUGAUUUUUAAUAAAUUGCCAUUUG .......(((((-((---....(..((--((-.-.......)))).).)))----))))...((((................))))..... ( -13.29) >consensus GAUUUGCUGUUGCUGGCGUUGUCGCCGUCCA_UUGCGAUUACGUUGGUCGGCGGUGGCCAGCAGCAUUGAUUUUUAAUAAAUAGCAAUUUG ...((((((((((((((....((((((.(((...(((....)))))).))))))..))))))))))...((((.....)))).)))).... (-17.92 = -21.48 + 3.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:48 2006