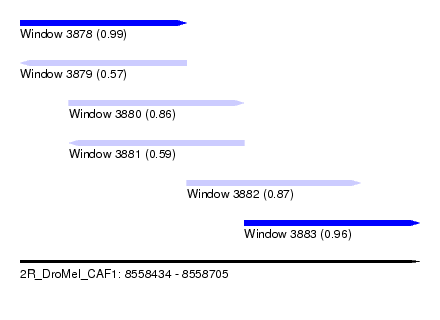
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,558,434 – 8,558,705 |
| Length | 271 |
| Max. P | 0.993472 |

| Location | 8,558,434 – 8,558,547 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -28.58 |
| Energy contribution | -30.26 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.26 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8558434 113 + 20766785 --GAAAAUGGUCAGGUAA-----UUGAAAAGCCAGCAGAGAGCUCAACUUGAUGCCAGCUCAAUUGGUCAACAGUACAGUCGUGCUUGACCGAUCUGCGUUGCUAUUAAAGCCUGGCCUG --......((((((((..-----...((.(((..((((((((((((......))..)))))..(((((((..(((((....)))))))))))))))))...))).))...)))))))).. ( -43.00) >DroSec_CAF1 34107 111 + 1 --UAAAAUGGUCAGGUAA-----UUGAAAAGGCAGCAGAGAGCUCGACUUAAUGGCAGCUCAAUUGGUCA--AGUACAGUUGUGCUGGGCCGAUCUGCGCUGCUUUUAAAGCCUGGCCUG --......((((((((..-----((((((.(((.((((((((((.(.((....))))))))..((((((.--(((((....))))).))))))))))))))..)))))).)))))))).. ( -48.00) >DroSim_CAF1 37411 111 + 1 --UAAAAUGGUCAGGUAA-----UUGAAAAGCCAGCGGAGAGCUCGACUUAAUGUCAGCUCAAUUGGUCA--AGUACAGUUGUGCUGGGCCGAUCUGCGCUGCUAUUAAAGCCUGGCCUG --......((((((((..-----...((.(((..((((((((((.(((.....))))))))..((((((.--(((((....))))).)))))))))))...))).))...)))))))).. ( -48.20) >DroEre_CAF1 22824 113 + 1 AUGAAAAUGGGCAGGUAA-----UUGAAAAGCCAGCUGAGAGCUGCACUUAAUGCAGGCUCUAUGGCUCC--AGUACAGUUGUGCUUGGCCGAACUGAGCUGCUAUUAAAGCCUGGCCUG ........((.(((((..-----...((.(((.((((.(((((((((.....)))).))))).(((((..--(((((....))))).))))).....))))))).))...))))).)).. ( -41.70) >DroYak_CAF1 37139 116 + 1 --GAAAAUGGACAGGUAAUGAAUUUGAAAUGCCAGCAGAGAGCUGGGCUUAAUGCCGGCUCUGUGGCUCA--AGUACAGUUGUGCCUGGCCGACCUGAGCUGCGAUUAAAGCCAGGUCUG --......((.((((((.((.((((((...((((....((((((((.(.....))))))))).)))))))--))).))....)))))).))((((((.(((........))))))))).. ( -49.10) >consensus __GAAAAUGGUCAGGUAA_____UUGAAAAGCCAGCAGAGAGCUCGACUUAAUGCCAGCUCAAUUGGUCA__AGUACAGUUGUGCUUGGCCGAUCUGCGCUGCUAUUAAAGCCUGGCCUG ........((((((((.................(((.....)))....((((((.((((.(((((((((...(((((....))))).))))))).)).)))).)))))).)))))))).. (-28.58 = -30.26 + 1.68)

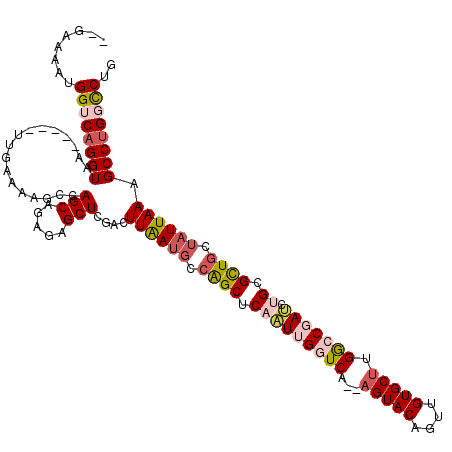

| Location | 8,558,434 – 8,558,547 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.34 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8558434 113 - 20766785 CAGGCCAGGCUUUAAUAGCAACGCAGAUCGGUCAAGCACGACUGUACUGUUGACCAAUUGAGCUGGCAUCAAGUUGAGCUCUCUGCUGGCUUUUCAA-----UUACCUGACCAUUUUC-- ..((.((((.......(((...(((((..(((((((((....)))....))))))....((((((((.....))).))))))))))..)))......-----...)))).))......-- ( -32.99) >DroSec_CAF1 34107 111 - 1 CAGGCCAGGCUUUAAAAGCAGCGCAGAUCGGCCCAGCACAACUGUACU--UGACCAAUUGAGCUGCCAUUAAGUCGAGCUCUCUGCUGCCUUUUCAA-----UUACCUGACCAUUUUA-- ..((.((((..((((((((((((.((((((((.((((.(((.((....--....)).))).)))).......)))))..))).))))).))))).))-----...)))).))......-- ( -30.30) >DroSim_CAF1 37411 111 - 1 CAGGCCAGGCUUUAAUAGCAGCGCAGAUCGGCCCAGCACAACUGUACU--UGACCAAUUGAGCUGACAUUAAGUCGAGCUCUCCGCUGGCUUUUCAA-----UUACCUGACCAUUUUA-- ..((.((((((.....)))......((..((((.(((.(((......)--)).......((((((((.....))).)))))...)))))))..))..-----....))).))......-- ( -30.40) >DroEre_CAF1 22824 113 - 1 CAGGCCAGGCUUUAAUAGCAGCUCAGUUCGGCCAAGCACAACUGUACU--GGAGCCAUAGAGCCUGCAUUAAGUGCAGCUCUCAGCUGGCUUUUCAA-----UUACCUGCCCAUUUUCAU ..((.((((.........(((..(((((..((...))..)))))..))--)((((((.(((((.(((((...))))))))))....)))))).....-----...)))).))........ ( -35.70) >DroYak_CAF1 37139 116 - 1 CAGACCUGGCUUUAAUCGCAGCUCAGGUCGGCCAGGCACAACUGUACU--UGAGCCACAGAGCCGGCAUUAAGCCCAGCUCUCUGCUGGCAUUUCAAAUUCAUUACCUGUCCAUUUUC-- ..(((((((((........))).))))))((.((((..(((......)--)).(((((((((..(((.....))).....))))).))))...............)))).))......-- ( -38.30) >consensus CAGGCCAGGCUUUAAUAGCAGCGCAGAUCGGCCAAGCACAACUGUACU__UGACCAAUUGAGCUGGCAUUAAGUCGAGCUCUCUGCUGGCUUUUCAA_____UUACCUGACCAUUUUC__ ..((.((((((.....)))......((..((((.(((((....))..............((((((((.....))).)))))...)))))))..))...........))).))........ (-19.62 = -20.34 + 0.72)

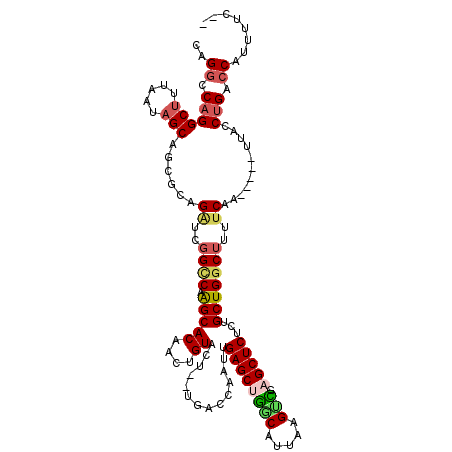
| Location | 8,558,467 – 8,558,586 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -40.74 |
| Consensus MFE | -26.16 |
| Energy contribution | -27.24 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8558467 119 + 20766785 AGCUCAACUUGAUGCCAGCUCAAUUGGUCAACAGUACAGUCGUGCUUGACCGAUCUGCGUUGCUAUUAAAGCCUGGCCUGG-AAGCCAAGUCUUUUUGCAUUCAAGGCCAAACAGAAACA .(((....((((((.((((.((((((((((..(((((....))))))))))))).)).)))).))))))))).((((((.(-((..((((....))))..))).)))))).......... ( -38.50) >DroSec_CAF1 34140 117 + 1 AGCUCGACUUAAUGGCAGCUCAAUUGGUCA--AGUACAGUUGUGCUGGGCCGAUCUGCGCUGCUUUUAAAGCCUGGCCUGG-AAGCCAAGGCUUUCUGCACUCAAGGCCAAACAAAAACC .(((....((((.((((((.(((((((((.--(((((....))))).))))))).)).)))))).))))))).((((((((-((((....))))))........)))))).......... ( -45.50) >DroSim_CAF1 37444 117 + 1 AGCUCGACUUAAUGUCAGCUCAAUUGGUCA--AGUACAGUUGUGCUGGGCCGAUCUGCGCUGCUAUUAAAGCCUGGCCUGG-AAGCCAAGUCUUUCUGCACUCAAGGCCAAACAAAAACC .(((....((((((.((((.(((((((((.--(((((....))))).))))))).)).)))).))))))))).((((((.(-(.((..((.....))))..)).)))))).......... ( -37.80) >DroEre_CAF1 22859 118 + 1 AGCUGCACUUAAUGCAGGCUCUAUGGCUCC--AGUACAGUUGUGCUUGGCCGAACUGAGCUGCUAUUAAAGCCUGGCCUGGUAAGCCAAGUCUUACUGCAUUCAAGGCCAAACAGAAACC ....((.((((((((((...((.(((((((--((..(((....(((..(.....)..))).(((.....))))))..))))..))))))).....))))))).)))))............ ( -36.90) >DroYak_CAF1 37177 118 + 1 AGCUGGGCUUAAUGCCGGCUCUGUGGCUCA--AGUACAGUUGUGCCUGGCCGACCUGAGCUGCGAUUAAAGCCAGGUCUGCAAUGCCAAGUCUUUUUGCAUUCAAGGCCAAAGCGAAACC .(((.((((((((((.(((..((((((.((--.((((....)))).)))))((((((.(((........))))))))).)))..)))(((....))))))))..)))))..)))...... ( -45.00) >consensus AGCUCGACUUAAUGCCAGCUCAAUUGGUCA__AGUACAGUUGUGCUUGGCCGAUCUGCGCUGCUAUUAAAGCCUGGCCUGG_AAGCCAAGUCUUUCUGCAUUCAAGGCCAAACAGAAACC .(((....((((((.((((.(((((((((...(((((....))))).))))))).)).)))).)))))))))..(((((.....((...........)).....)))))........... (-26.16 = -27.24 + 1.08)

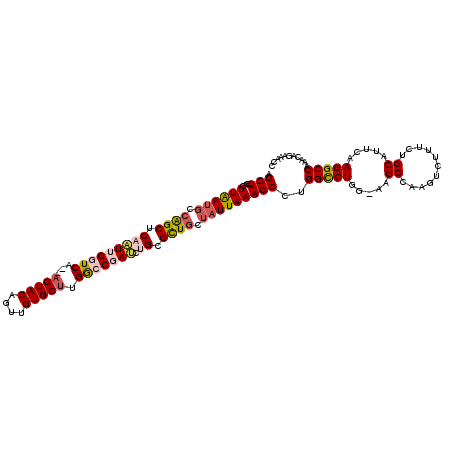

| Location | 8,558,467 – 8,558,586 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -39.64 |
| Consensus MFE | -27.76 |
| Energy contribution | -29.00 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.590920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8558467 119 - 20766785 UGUUUCUGUUUGGCCUUGAAUGCAAAAAGACUUGGCUU-CCAGGCCAGGCUUUAAUAGCAACGCAGAUCGGUCAAGCACGACUGUACUGUUGACCAAUUGAGCUGGCAUCAAGUUGAGCU .......(((..((.((((.(((..((((.(((((((.-...)))))))))))..((((...((((..(((((......)))))..))))....(....).)))))))))))))..))). ( -37.20) >DroSec_CAF1 34140 117 - 1 GGUUUUUGUUUGGCCUUGAGUGCAGAAAGCCUUGGCUU-CCAGGCCAGGCUUUAAAAGCAGCGCAGAUCGGCCCAGCACAACUGUACU--UGACCAAUUGAGCUGCCAUUAAGUCGAGCU (((.(((..((((..(..((((((((((((((.((((.-...)))))))))))....((.(.((......)).).))....)))))))--..)))))..)))..)))............. ( -38.10) >DroSim_CAF1 37444 117 - 1 GGUUUUUGUUUGGCCUUGAGUGCAGAAAGACUUGGCUU-CCAGGCCAGGCUUUAAUAGCAGCGCAGAUCGGCCCAGCACAACUGUACU--UGACCAAUUGAGCUGACAUUAAGUCGAGCU .........((((..(..(((((((((((.(((((((.-...)))))))))))....((.(.((......)).).))....)))))))--..)))))...(((((((.....))).)))) ( -37.30) >DroEre_CAF1 22859 118 - 1 GGUUUCUGUUUGGCCUUGAAUGCAGUAAGACUUGGCUUACCAGGCCAGGCUUUAAUAGCAGCUCAGUUCGGCCAAGCACAACUGUACU--GGAGCCAUAGAGCCUGCAUUAAGUGCAGCU ((((((.((((((((((((.(((..(((..(((((((.....)))))))..)))...)))..))))...))))))))((....))...--))))))....(((.(((((...)))))))) ( -44.00) >DroYak_CAF1 37177 118 - 1 GGUUUCGCUUUGGCCUUGAAUGCAAAAAGACUUGGCAUUGCAGACCUGGCUUUAAUCGCAGCUCAGGUCGGCCAGGCACAACUGUACU--UGAGCCACAGAGCCGGCAUUAAGCCCAGCU ......((((.((((.(((((((...........))))).))(((((((((........))).))))))))))))))....(((..((--((((((........))).)))))..))).. ( -41.60) >consensus GGUUUCUGUUUGGCCUUGAAUGCAGAAAGACUUGGCUU_CCAGGCCAGGCUUUAAUAGCAGCGCAGAUCGGCCAAGCACAACUGUACU__UGACCAAUUGAGCUGGCAUUAAGUCGAGCU ((((..((((((((((((..(((..((((.(((((((.....)))))))))))....)))...)))...))))))))).)))).................(((((((.....))).)))) (-27.76 = -29.00 + 1.24)

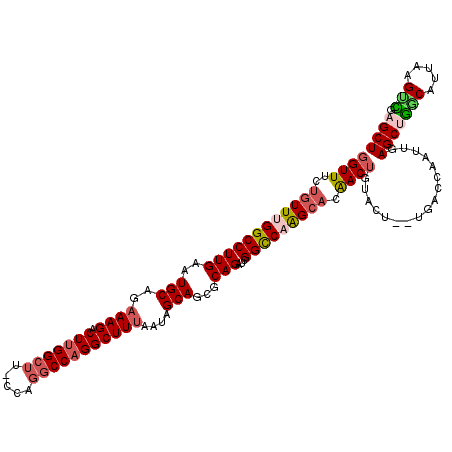
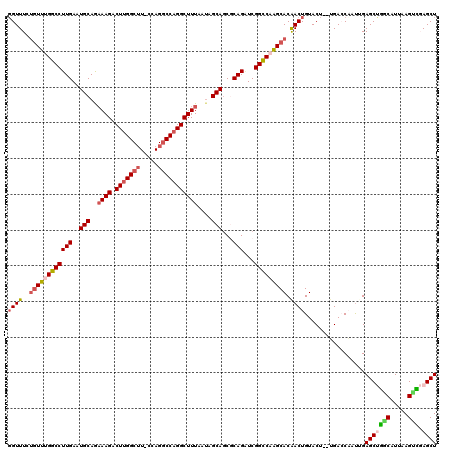
| Location | 8,558,547 – 8,558,665 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8558547 118 + 20766785 G-AAGCCAAGUCUUUUUGCAUUCAAGGCCAAACAGAAACAUACACGUCUUCCCUUCAUUAUUCAUUU-GCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUA (-((...((((...(((((......(......).............((((((((((((..(((....-..))).)))).))))))))...)))))..))))...)))............. ( -22.80) >DroSec_CAF1 34218 118 + 1 G-AAGCCAAGGCUUUCUGCACUCAAGGCCAAACAAAAACCUACACGUCUUCCCUUCAUUAUUCAUUU-GCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUA (-(((....((((((........)))))).................((((((((((((..(((....-..))).)))).))))))))...........)))).................. ( -24.30) >DroSim_CAF1 37522 118 + 1 G-AAGCCAAGUCUUUCUGCACUCAAGGCCAAACAAAAACCUACACGUCUUCCCUUCAUUAUUCAUUU-GCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUA (-((...((((.....(((.....(((...........))).....((((((((((((..(((....-..))).)))).))))))))...)))....))))...)))............. ( -21.90) >DroEre_CAF1 22937 119 + 1 GUAAGCCAAGUCUUACUGCAUUCAAGGCCAAACAGAAACCAACACGUCUUCCCUUCAUUAUUCAUUU-GCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUA (((((......))))).........(((..................((((((((((((..(((....-..))).)))).))))))))..........((((.......))))..)))... ( -22.30) >DroYak_CAF1 37255 120 + 1 CAAUGCCAAGUCUUUUUGCAUUCAAGGCCAAAGCGAAACCUACACGUCUUCCCUUCAUUAUUCAUUUGGCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUA .(((((.(((....))))))))...(((....((............((((((((((((..(((.......))).)))).))))))))...)).....((((.......))))..)))... ( -22.06) >consensus G_AAGCCAAGUCUUUCUGCAUUCAAGGCCAAACAGAAACCUACACGUCUUCCCUUCAUUAUUCAUUU_GCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUA .........................(((..................((((((((((((..(((.......))).)))).))))))))..........((((.......))))..)))... (-19.00 = -19.00 + 0.00)

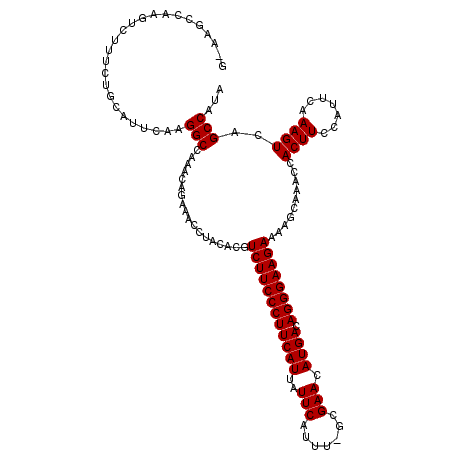
| Location | 8,558,586 – 8,558,705 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -23.70 |
| Energy contribution | -23.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8558586 119 + 20766785 UACACGUCUUCCCUUCAUUAUUCAUUU-GCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUAUGGAAAGAUAUCUUUUCAAACAAAGUGAGCAAAGAGUAGU ................(((((((.(((-((..((.((((..(((((............))))).......))))......(((((((....)))))))......))..)))))))))))) ( -25.10) >DroSec_CAF1 34257 119 + 1 UACACGUCUUCCCUUCAUUAUUCAUUU-GCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUAUGGAAAGAUAUCUUUUCAAACAAAGUGAGCAAAGAGUAGU ................(((((((.(((-((..((.((((..(((((............))))).......))))......(((((((....)))))))......))..)))))))))))) ( -25.10) >DroSim_CAF1 37561 119 + 1 UACACGUCUUCCCUUCAUUAUUCAUUU-GCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUAUGGAAAGAUAUCUUUUCAAACAAAGUGAGCAAAGAGUAGU ................(((((((.(((-((..((.((((..(((((............))))).......))))......(((((((....)))))))......))..)))))))))))) ( -25.10) >DroEre_CAF1 22977 119 + 1 AACACGUCUUCCCUUCAUUAUUCAUUU-GCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUAUGGAAAGAUAUCUUUUCAAACAAAGUGAGCAAAGAGUAGU ................(((((((.(((-((..((.((((..(((((............))))).......))))......(((((((....)))))))......))..)))))))))))) ( -25.10) >DroYak_CAF1 37295 120 + 1 UACACGUCUUCCCUUCAUUAUUCAUUUGGCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUAUGGAAAGAUAUCUUUUCAAACAAAGUGAGCAAAGAGUAGU ...((.((((..((.((((.......((((((...(((...(((((............)))))..)))...)).))))..(((((((....))))))).....))))))..))))))... ( -25.10) >consensus UACACGUCUUCCCUUCAUUAUUCAUUU_GCGAACAUGACAGGGAAGAAAAGCAAACCACUUCCAUUCAAAGUCAGCCAUAUGGAAAGAUAUCUUUUCAAACAAAGUGAGCAAAGAGUAGU ..(((.((((((((((((..(((.......))).)))).)))))))).................................(((((((....)))))))......)))............. (-23.70 = -23.70 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:45 2006