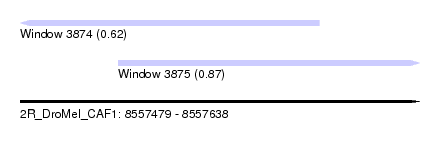
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,557,479 – 8,557,638 |
| Length | 159 |
| Max. P | 0.874853 |

| Location | 8,557,479 – 8,557,598 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -22.68 |
| Energy contribution | -23.12 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
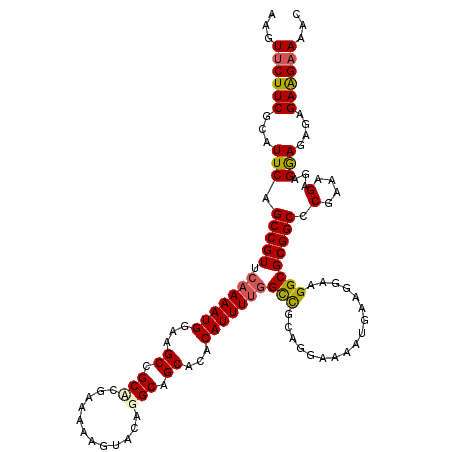
>2R_DroMel_CAF1 8557479 119 - 20766785 CAUUUUCCUGCGACCAAAAUGUGUGCUGCCUGUACUUUUUCAGGCGGCUUCCAUUUUGAACGGCUGAAUGCGAAGAACUUCUGUUUUGAAAAAUGUAUCUCCUUCGCAGUACGUAGA-AU .......((((((((((((((...((((((((........))))))))...)))))))..........((((((((((....)))((....))........))))))))).))))).-.. ( -37.10) >DroSec_CAF1 33169 119 - 1 UAUUUUCCUGCGGCCAAAAUGUGUGCUGCCUGUACUUUUUCGUGCGGCUUCCAUUUUGAACGGCUGAAUGCGAAGAACUGCUGUUUUGGAAAAUGUAUCUCUUAAGCAGUACACAGA-AU ...................((((((((((((((((......)))))).(((((....(((((((...............))))))))))))..............))))))))))..-.. ( -33.46) >DroSim_CAF1 36463 119 - 1 CAUUUUCCUGCGGCCAAAAUGUGUGCUGCCUGUACUUUUUCGUGCGGCUUCCAUUUUGAACGGCUGAAUGCGAAGAACUGCUGUUUUGGAAAAUGUAUCUCUUAAGCAGUACACAGA-AU ...................((((((((((((((((......)))))).(((((....(((((((...............))))))))))))..............))))))))))..-.. ( -33.46) >DroEre_CAF1 21960 110 - 1 CAUUUUCCUACGGCCGAAAUG--UGCUGCCUGUACUUUUUGGAGCGGCUUCCAUUUUGAACGGCUGAAUGCGAAGAACUUCU-UUUUGGGAAAUGUAUAUCUUU-------UUCAAAUUC ((((((((..(((((((((((--.((((((((.......))).)))))...)))))....)))))).....((((.....))-))..)))))))).........-------......... ( -28.90) >DroYak_CAF1 36273 116 - 1 CAUUUUCCUCUAGCAAAAAUG--CGCUGCCUGUACUUUUUGCAGCGGCUUCCAUUUUGAACGGCUGAAUGCGAAGAACUUCU-UUUUGGAAAAUGUAUCUCUUCCGCGGAAUUCAGA-UC ((((((((..((((.((((((--.(((((.((((.....)))))))))...)))))).....)))).....((((.....))-))..)))))))).((((.(((....)))...)))-). ( -27.80) >consensus CAUUUUCCUGCGGCCAAAAUGUGUGCUGCCUGUACUUUUUCGAGCGGCUUCCAUUUUGAACGGCUGAAUGCGAAGAACUUCUGUUUUGGAAAAUGUAUCUCUUAAGCAGUACACAGA_AU ((((((((..((..(((((((...(((((.((........)).)))))...)))))))..))((.....))................))))))))......................... (-22.68 = -23.12 + 0.44)


| Location | 8,557,518 – 8,557,638 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -27.19 |
| Energy contribution | -27.47 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8557518 120 + 20766785 AAGUUCUUCGCAUUCAGCCGUUCAAAAUGGAAGCCGCCUGAAAAAGUACAGGCAGCACACAUUUUGGUCGCAGGAAAAUGAAGGAAGGCGCGGCCCGAAAGAGAGGAGAGAGAAGAAGAC ...((((((.(.(((.(((((.(((((((...((.(((((........))))).))...)))))))(((.((......))......)))))))).(....)....))).).))))))... ( -36.10) >DroSec_CAF1 33208 120 + 1 CAGUUCUUCGCAUUCAGCCGUUCAAAAUGGAAGCCGCACGAAAAAGUACAGGCAGCACACAUUUUGGCCGCAGGAAAAUAAAGGAAGGCGCGGCCCGAAAGAGAGGAGAGAGAAGAACAC ..(((((((.(.(((.(((((.(((((((...(((..((......))...)))......)))))))(((.................)))))))).(....)....))).).))))))).. ( -34.73) >DroSim_CAF1 36502 120 + 1 CAGUUCUUCGCAUUCAGCCGUUCAAAAUGGAAGCCGCACGAAAAAGUACAGGCAGCACACAUUUUGGCCGCAGGAAAAUGAAGGAAGGCGCGGCCCGAAAGAGAGGAGAGAGAAGAACAC ..(((((((.(.(((.(((((.(((((((...(((..((......))...)))......)))))))(((.((......))......)))))))).(....)....))).).))))))).. ( -34.90) >DroEre_CAF1 21992 118 + 1 AAGUUCUUCGCAUUCAGCCGUUCAAAAUGGAAGCCGCUCCAAAAAGUACAGGCAGCA--CAUUUCGGCCGUAGGAAAAUGAAGGAAGGCGCGGCCCGAAAGAGAGGAGAGAGAAGAAAAC ...((((((.(.(((.(((((......((((......))))......)).)))....--..((((((((((..................)))).)))))).....))).).))))))... ( -28.67) >DroYak_CAF1 36311 118 + 1 AAGUUCUUCGCAUUCAGCCGUUCAAAAUGGAAGCCGCUGCAAAAAGUACAGGCAGCG--CAUUUUUGCUAGAGGAAAAUGAAGGAGGGCGCGGCGCGAAAGAGAGAAGAGAGAGGUAAAC ...((((((.(.(((.(((((((......)).((((((((...........))))).--(((((((.......)))))))......))))))))..))).)...)))))).......... ( -32.20) >consensus AAGUUCUUCGCAUUCAGCCGUUCAAAAUGGAAGCCGCACGAAAAAGUACAGGCAGCACACAUUUUGGCCGCAGGAAAAUGAAGGAAGGCGCGGCCCGAAAGAGAGGAGAGAGAAGAAAAC ...((((((...(((.(((((.(((((((...((.(((............))).))...)))))))(((.................)))))))).(....)...)))....))))))... (-27.19 = -27.47 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:37 2006