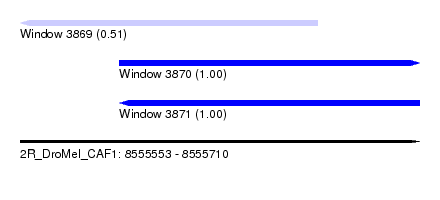
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,555,553 – 8,555,710 |
| Length | 157 |
| Max. P | 0.999232 |

| Location | 8,555,553 – 8,555,670 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.56 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -21.85 |
| Energy contribution | -21.97 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8555553 117 - 20766785 A--AGAUAUCGUGCUGUGAAUGAAAUUGCCAUCUAAAGUGGCUUUUGCUAUCGAUUUCGAUAGCUGGCAUCAAUUGCCGCUCGAUGAAACCGAAAUGCGAC-CCGUCGACCGCAGAAAAU .--...(((((.((.........((..(((((.....)))))..))(((((((....))))))).((((.....)))))).))))).........((((..-........))))...... ( -33.70) >DroSec_CAF1 31182 118 - 1 A-AAGAUAUCGUGCUGUGAAUGAAAUUCCCAUCUAAAGUGGUUUUUGCUAUCGAUUUCGAUAGCUGGCAUCAAUAGCCGCUCGAUGAAACCGAAAUGCGAC-CCGUCGACCGCAGAAAAU .-...........(((((..(((...((.(((......(((((((.(((((((....))))))).(((.......))).......)))))))..))).)).-...)))..)))))..... ( -28.50) >DroSim_CAF1 34755 118 - 1 A-AAGAUAUCGUGCUCUGAAUGAAAUUCCCAUCUAAAGUGGUUUUUGCUAUCGAUUUCGAUAGCUGGCAUCAAUUGCCGCUCGAUGAAACUGAAAUGCGAC-CCGUCGACCGCAGAAAAU .-....(((((.((..............((((.....)))).....(((((((....))))))).((((.....)))))).)))))...(((.....(((.-...)))....)))..... ( -32.20) >DroEre_CAF1 20079 119 - 1 UAAUGAUAUAGUGCCAUGAAUGAAAUA-CCAUAUAUGGCGUCAUUGGCUAUCGAUUUCGAUAGCUGGCAUCGAUUGCCGCUCGAUGAAACCGAAAUGCGACCCCGUCGACCGCAGAAAAU .((((((.....((((((.(((.....-.))).))))))))))))((((((((....))))))))((((.....))))((((((((....((.....))....))))))..))....... ( -37.90) >DroYak_CAF1 34429 101 - 1 ----------AUACAAUGAAUGAAAUU-CCAUAUAUAGUGAAACUGGCUAUCGAUUUCGAUUGCUGACGUCGAUUGCCGCUCGAUGAAACCGAA--------CCGUCGACCGCAGAAAAU ----------.(((.(((.(((.....-.))).))).))).....(((.((((((.(((.....))).)))))).)))((((((((........--------.))))))..))....... ( -23.60) >consensus A_AAGAUAUCGUGCUAUGAAUGAAAUU_CCAUCUAAAGUGGCUUUUGCUAUCGAUUUCGAUAGCUGGCAUCAAUUGCCGCUCGAUGAAACCGAAAUGCGAC_CCGUCGACCGCAGAAAAU ...........((((.((.(((.......))).)).))))......(((((((....))))))).((((.....))))((((((((.................))))))..))....... (-21.85 = -21.97 + 0.12)

| Location | 8,555,592 – 8,555,710 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.98 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8555592 118 + 20766785 GCGGCAAUUGAUGCCAGCUAUCGAAAUCGAUAGCAAAAGCCACUUUAGAUGGCAAUUUCAUUCACAGCACGAUAUCU--UUACAACUGAAAGUUAAAAUAAAGCGGUAAAUAAAUACAAA ((((((.....)))).(((((((....)))))))....(((((....).)))).............))........(--((((..((..............))..))))).......... ( -26.64) >DroSec_CAF1 31221 119 + 1 GCGGCUAUUGAUGCCAGCUAUCGAAAUCGAUAGCAAAAACCACUUUAGAUGGGAAUUUCAUUCACAGCACGAUAUCUU-UUACAACUGAAACUUAAAAUAAAGCGCUAAAUAAAUAAGAA (((((.......))).(((((((....))))))).........(((((.((..((......((.......)).....)-)..)).)))))............))................ ( -21.30) >DroSim_CAF1 34794 114 + 1 GCGGCAAUUGAUGCCAGCUAUCGAAAUCGAUAGCAAAAACCACUUUAGAUGGGAAUUUCAUUCAGAGCACGAUAUCUU-UUACAACUAAAUGUUA---UUCAGCG--GAAUAAAUAAAAA ((((((.....)))).(((((((....))))))).....((((....).)))((((..((((.((.............-......)).))))..)---))).)).--............. ( -24.11) >DroEre_CAF1 20119 108 + 1 GCGGCAAUCGAUGCCAGCUAUCGAAAUCGAUAGCCAAUGACGCCAUAUAUGG-UAUUUCAUUCAUGGCACUAUAUCAUUAUAUAACUAAAUGCUUAGAUAAAA----------AGAAU-A ((((((.....)))).(((((((....))))))).......(((((..((((-....))))..))))).......................))..........----------.....-. ( -27.20) >DroYak_CAF1 34461 95 + 1 GCGGCAAUCGACGUCAGCAAUCGAAAUCGAUAGCCAGUUUCACUAUAUAUGG-AAUUUCAUUCAUUGUAU-------------AACAAAAUGCUAAAAUAAAA----------AUAAU-A (((((.(((((..((.......))..))))).)))........(((((((((-(......)))).)))))-------------).......))..........----------.....-. ( -17.90) >consensus GCGGCAAUUGAUGCCAGCUAUCGAAAUCGAUAGCAAAAACCACUUUAGAUGG_AAUUUCAUUCACAGCACGAUAUCUU_UUACAACUAAAUGUUAAAAUAAAGCG___AAUAAAUAAAAA ..(((.......))).(((((((....)))))))........((((..((((.....))))..))))..................................................... (-16.28 = -16.52 + 0.24)

| Location | 8,555,592 – 8,555,710 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.98 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.44 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.53 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8555592 118 - 20766785 UUUGUAUUUAUUUACCGCUUUAUUUUAACUUUCAGUUGUAA--AGAUAUCGUGCUGUGAAUGAAAUUGCCAUCUAAAGUGGCUUUUGCUAUCGAUUUCGAUAGCUGGCAUCAAUUGCCGC ......(((((((((.((..((((((.((........)).)--)))))....)).)))))))))...(((((.....)))))....(((((((....))))))).((((.....)))).. ( -35.50) >DroSec_CAF1 31221 119 - 1 UUCUUAUUUAUUUAGCGCUUUAUUUUAAGUUUCAGUUGUAA-AAGAUAUCGUGCUGUGAAUGAAAUUCCCAUCUAAAGUGGUUUUUGCUAUCGAUUUCGAUAGCUGGCAUCAAUAGCCGC ...((((((((..(((((..((((((...............-))))))..))))))))))))).....((((.....)))).....(((((((....))))))).(((.......))).. ( -28.46) >DroSim_CAF1 34794 114 - 1 UUUUUAUUUAUUC--CGCUGAA---UAACAUUUAGUUGUAA-AAGAUAUCGUGCUCUGAAUGAAAUUCCCAUCUAAAGUGGUUUUUGCUAUCGAUUUCGAUAGCUGGCAUCAAUUGCCGC ............(--(((((((---(..(((((((..(((.-..(....).))).)))))))..))))........))))).....(((((((....))))))).((((.....)))).. ( -29.60) >DroEre_CAF1 20119 108 - 1 U-AUUCU----------UUUUAUCUAAGCAUUUAGUUAUAUAAUGAUAUAGUGCCAUGAAUGAAAUA-CCAUAUAUGGCGUCAUUGGCUAUCGAUUUCGAUAGCUGGCAUCGAUUGCCGC .-.....----------..........((............((((((.....((((((.(((.....-.))).))))))))))))((((((((....))))))))((((.....)))))) ( -30.60) >DroYak_CAF1 34461 95 - 1 U-AUUAU----------UUUUAUUUUAGCAUUUUGUU-------------AUACAAUGAAUGAAAUU-CCAUAUAUAGUGAAACUGGCUAUCGAUUUCGAUUGCUGACGUCGAUUGCCGC .-.....----------..........((.....(((-------------.(((.(((.(((.....-.))).))).))).))).(((.((((((.(((.....))).)))))).))))) ( -19.40) >consensus UUAUUAUUUAUU___CGCUUUAUUUUAACAUUUAGUUGUAA_AAGAUAUCGUGCUAUGAAUGAAAUU_CCAUCUAAAGUGGCUUUUGCUAUCGAUUUCGAUAGCUGGCAUCAAUUGCCGC ...................................................((((.((.(((.......))).)).))))......(((((((....))))))).((((.....)))).. (-15.32 = -15.44 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:33 2006