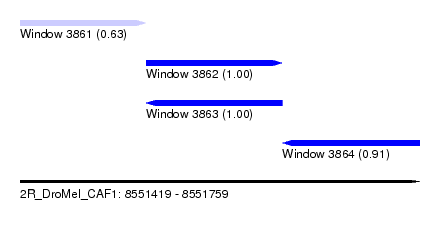
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,551,419 – 8,551,759 |
| Length | 340 |
| Max. P | 0.999812 |

| Location | 8,551,419 – 8,551,526 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.49 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -22.44 |
| Energy contribution | -22.20 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8551419 107 + 20766785 CUUCGUGUCGUCGUGGCUCGGCCUCUUUCAUCAGCAUAUUUCUGAGUGAUGAAAAUUAUGUUGAUGACGUUUGCACAGCGCCCAG---UGCCCCAACCCACC----------CCACAACA .........((.((((...((......(((((((((((((((........))))..))))))))))).....((((........)---)))......))...----------)))).)). ( -27.20) >DroSec_CAF1 26947 110 + 1 CUUCGUGUCGUCGUGGCUCGGCCUCUUUCAUCAGCAUAUUUCUGAGUGAUGAAAAUUAUGUUGAUGACGAUUGCACAGCGCCCAGUAGUACCCCAACCCACC----------CCACGACA .........(((((((((.(((.....(((((((((((((((........))))..))))))))))).....((...))))).))..((......)).....----------))))))). ( -30.10) >DroSim_CAF1 30587 110 + 1 CUUCGUGUCGUCGUGGCUCGGCCUCUUUCAUCAGCAUAUUUCUGAGUGAUGAAAAUUAUGUUGAUGACGUUUGCACAGCGCCCAGUAGUACCCCAACCCACC----------CCACGACA .........(((((((((.(((.....(((((((((((((((........))))..))))))))))).(((.....)))))).))..((......)).....----------))))))). ( -30.30) >DroEre_CAF1 15988 105 + 1 CUUCGUGUCGUCGUGGUUCGGCCUCUUUCAUCAGCAUAUUUCUGAGUGAUGAAAAUUAUGUUGAUGACGUUUGCACAGCGCCCGU---UACCCCACCUCACGACCC------------CU ..(((((..((.(.(((.(((......(((((((((((((((........))))..)))))))))))((((.....)))).))).---.)))).))..)))))...------------.. ( -30.90) >DroYak_CAF1 30212 117 + 1 CUUCGUGUCGUCGUGGUUCGGCCUCUUUCAUCAGCAUAUUUCUGAGUGAUGAAAAUUAUGUUGAUGACGUUUGCACAGCGCCCGU---UACCCCACCCCACCGCACCACAUCCCACAACC ....(((..((.(((((.(((......(((((((((((((((........))))..)))))))))))((..(((...)))..)).---............))).)))))))..))).... ( -32.50) >consensus CUUCGUGUCGUCGUGGCUCGGCCUCUUUCAUCAGCAUAUUUCUGAGUGAUGAAAAUUAUGUUGAUGACGUUUGCACAGCGCCCAG___UACCCCAACCCACC__________CCACAACA ....((((...((((((...))).....((((((((((((((........))))..)))))))))))))...))))............................................ (-22.44 = -22.20 + -0.24)

| Location | 8,551,526 – 8,551,642 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -27.66 |
| Energy contribution | -27.70 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8551526 116 + 20766785 CCUC-CCCCUUCCAUAUUUUUAAGCCCCAGCGGCGACAACUAGCGAAAGUUGCGCUGAAUUGGAACAUCAGCGUUUUGCCGCUGCUUCUGUUUAUUUCUUUGAGCUUUUUCACUUU---U ....-......................(((((((((.....(((....)))(((((((.........))))))).))))))))).....(((((......)))))...........---. ( -29.70) >DroSec_CAF1 27057 116 + 1 CCUU-CCCCUUCCCCAUUUUUAAGCCCCAGCGGCGACAACUAGCGAAAGUUGCGCUGAAUUGGAACAUCAGCGUUUUGCCGUUGCUUCUGUUUAUUUCUUCGAGCUUUUUCACUUU---U ....-...............(((((..(((((((((.....(((....)))(((((((.........))))))).))))))))).....)))))......................---. ( -26.20) >DroSim_CAF1 30697 116 + 1 CCUU-CCCCUUCCCCAUUUUUAAGCCCCAGCGGCGACAACUAGCGAAAGUUGCGCUGAAUUGGAACAUCAGCGUUUUGCCGCUGCUUCUGUUUAUUUCUUCGAGCUUUUUCACUUU---U ....-...............(((((..(((((((((.....(((....)))(((((((.........))))))).))))))))).....)))))......................---. ( -28.60) >DroEre_CAF1 16093 120 + 1 CCUCCCCGCUUCCACAUUUUCAAGCCCCAGCGGCGACAACUAGCGAAAGUUGCGCUGAAUUGGAACAUCAGCGUUUUGCCGCUGCUUCUGUUUAUUUCUUCGAGCUUUUUCAUUUUCUCU .......((((...........((...(((((((((.....(((....)))(((((((.........))))))).)))))))))...))............))))............... ( -30.90) >DroYak_CAF1 30329 119 + 1 CCUCCCCCCUUUCUCAUUUUUAAGCCCCAGCGGCGACAACUAGCGAAAGUUGCGCUGAAUUGGAACAUCAGCGUUUUGCCGCUGCUUCUGUUUAUUUCUUUGAGCUUUUUCACUUU-ACU ............((((....(((((..(((((((((.....(((....)))(((((((.........))))))).))))))))).....)))))......))))............-... ( -31.90) >consensus CCUC_CCCCUUCCACAUUUUUAAGCCCCAGCGGCGACAACUAGCGAAAGUUGCGCUGAAUUGGAACAUCAGCGUUUUGCCGCUGCUUCUGUUUAUUUCUUCGAGCUUUUUCACUUU___U ....................(((((..(((((((((.....(((....)))(((((((.........))))))).))))))))).....))))).......................... (-27.66 = -27.70 + 0.04)


| Location | 8,551,526 – 8,551,642 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -31.70 |
| Energy contribution | -31.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8551526 116 - 20766785 A---AAAGUGAAAAAGCUCAAAGAAAUAAACAGAAGCAGCGGCAAAACGCUGAUGUUCCAAUUCAGCGCAACUUUCGCUAGUUGUCGCCGCUGGGGCUUAAAAAUAUGGAAGGGG-GAGG .---.........((((((..(....).........(((((((.....(((((.........)))))((((((......)))))).)))))))))))))................-.... ( -33.00) >DroSec_CAF1 27057 116 - 1 A---AAAGUGAAAAAGCUCGAAGAAAUAAACAGAAGCAACGGCAAAACGCUGAUGUUCCAAUUCAGCGCAACUUUCGCUAGUUGUCGCCGCUGGGGCUUAAAAAUGGGGAAGGGG-AAGG .---.........((((((..(....).........((.((((.....(((((.........)))))((((((......)))))).)))).))))))))................-.... ( -26.40) >DroSim_CAF1 30697 116 - 1 A---AAAGUGAAAAAGCUCGAAGAAAUAAACAGAAGCAGCGGCAAAACGCUGAUGUUCCAAUUCAGCGCAACUUUCGCUAGUUGUCGCCGCUGGGGCUUAAAAAUGGGGAAGGGG-AAGG .---.........((((((..(....).........(((((((.....(((((.........)))))((((((......)))))).)))))))))))))................-.... ( -33.00) >DroEre_CAF1 16093 120 - 1 AGAGAAAAUGAAAAAGCUCGAAGAAAUAAACAGAAGCAGCGGCAAAACGCUGAUGUUCCAAUUCAGCGCAACUUUCGCUAGUUGUCGCCGCUGGGGCUUGAAAAUGUGGAAGCGGGGAGG .............((((((..(....).........(((((((.....(((((.........)))))((((((......)))))).)))))))))))))..................... ( -33.00) >DroYak_CAF1 30329 119 - 1 AGU-AAAGUGAAAAAGCUCAAAGAAAUAAACAGAAGCAGCGGCAAAACGCUGAUGUUCCAAUUCAGCGCAACUUUCGCUAGUUGUCGCCGCUGGGGCUUAAAAAUGAGAAAGGGGGGAGG ...-.........((((((..(....).........(((((((.....(((((.........)))))((((((......)))))).)))))))))))))..................... ( -33.00) >consensus A___AAAGUGAAAAAGCUCGAAGAAAUAAACAGAAGCAGCGGCAAAACGCUGAUGUUCCAAUUCAGCGCAACUUUCGCUAGUUGUCGCCGCUGGGGCUUAAAAAUGGGGAAGGGG_GAGG .............((((((..(....).........(((((((.....(((((.........)))))((((((......)))))).)))))))))))))..................... (-31.70 = -31.90 + 0.20)

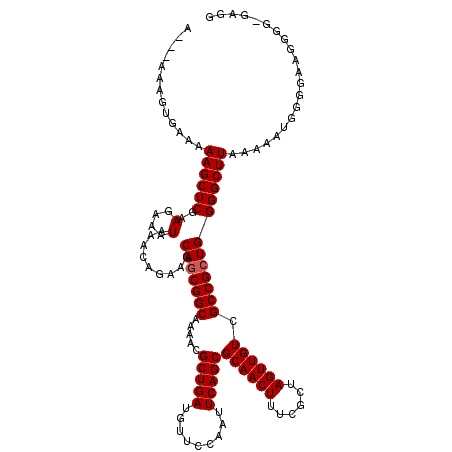
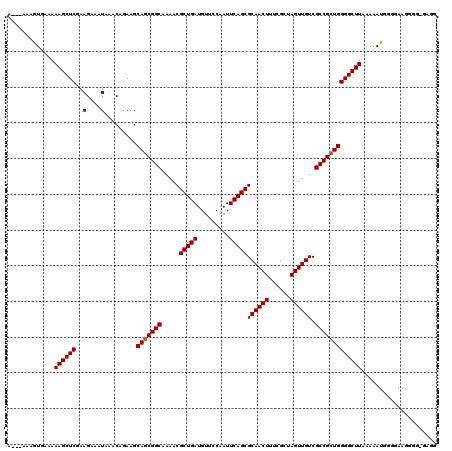
| Location | 8,551,642 – 8,551,759 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -37.84 |
| Consensus MFE | -28.18 |
| Energy contribution | -27.86 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8551642 117 - 20766785 CACAAAAAGUAGCCGCAAAAUACGCACAUAAAUUUUUGCGUUCAUGUAAAUGAAUUUUUCGAAUUCCGCUUUGCGUGGCUACUUUGCUUUUUGAUGUGGCGGC---GGCGGCGAAGAGAG .....(((((((((((.....(((((.(......).)))))....(((((.((((((...))))))...)))))))))))))))).((((((..(((.((...---.)).))).)))))) ( -33.80) >DroSec_CAF1 27173 116 - 1 CACAAAAAGUAGCCGCAAAAUACGCACAUAAAUU-UUGCGUUCAUGUAAAUGAAUUUUUCGAAUUCCGCUUUGCGUGGCUACUUUGCUUUUUGCUGUGGCGGC---GGCGGCGAAGAGAG .....(((((((((((.....(((((........-.)))))....(((((.((((((...))))))...)))))))))))))))).(((((((((((.(...)---.))))))))))).. ( -39.30) >DroSim_CAF1 30813 119 - 1 CACAAAAAGUAGCCGCAAAAUACGCACAUAAAUU-UUGCGUUCAUGUAAAUGAAUUUUUCGAAUUCCGCUUUGCGUGGCUACUUUGCUUUUUGCUGUGGCGGCGGCGGCGGCGAAGAGAG .....(((((((((((.....(((((........-.)))))....(((((.((((((...))))))...)))))))))))))))).(((((((((((.((....)).))))))))))).. ( -44.40) >DroEre_CAF1 16213 112 - 1 CACAAAAAGUAGCCGCAAAAUACGCACAUAAAUU-UUGCGUUCAUGUAAAUGAAUUUUUCGAGUUCCGCUUUGUGUGGCUACUUUGCU-------GCGGCGACGGCGGCGGCGAAGAGAG .....((((((((((((....(((((........-.)))))((((....))))....................))))))))))))(((-------((.((....)).)))))........ ( -37.00) >DroYak_CAF1 30448 109 - 1 CACAAAGAGUAGCCGCAAAAUACGCACAUAAAUU-UUGCGUUCAUGUAAAUGAAUUUUUCGAAUUCCGCUUUGCGUGGCUACUUUGCU-------GCGGCGACGGCG---GCAAAGAGAG .....(((((((((((.....(((((........-.)))))....(((((.((((((...))))))...))))))))))))))))(((-------(((....).)))---))........ ( -34.70) >consensus CACAAAAAGUAGCCGCAAAAUACGCACAUAAAUU_UUGCGUUCAUGUAAAUGAAUUUUUCGAAUUCCGCUUUGCGUGGCUACUUUGCUUUUUG_UGUGGCGGCGGCGGCGGCGAAGAGAG .....(((((((((((.....(((((..........)))))....(((((.((((((...))))))...))))))))))))))))..........((.((.......)).))........ (-28.18 = -27.86 + -0.32)

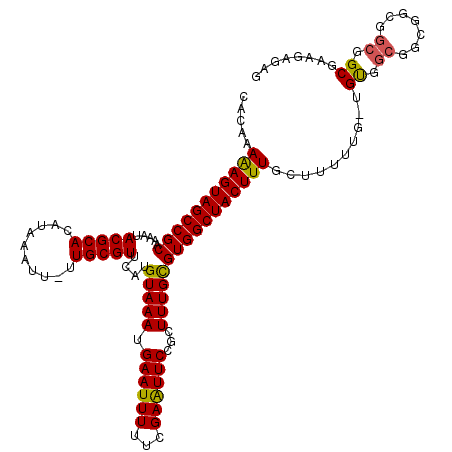
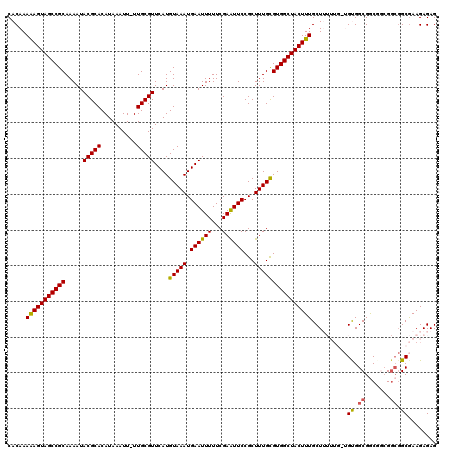
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:27 2006