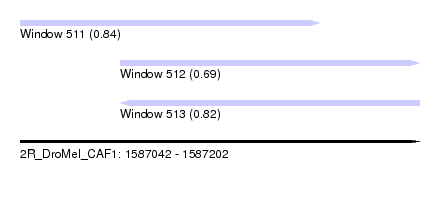
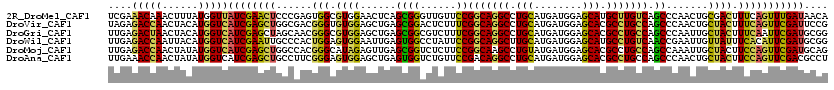
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,587,042 – 1,587,202 |
| Length | 160 |
| Max. P | 0.838984 |

| Location | 1,587,042 – 1,587,162 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.28 |
| Mean single sequence MFE | -46.93 |
| Consensus MFE | -27.67 |
| Energy contribution | -28.53 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1587042 120 + 20766785 CGCAAGUCACAGCCAGCACGCACAGCAACUUGCUGAGUCUUGUUAUCAAACUGAAAGUCGCAGUUGGGCUGACAAGCAUGCUCCAUCAUGCAGGCCUGCCGGAACAACCCGCUGAGUUCC .(((.(((.(((((((((.((.((((.....)))).))..))))....(((((.......))))).)))))....(((((......))))).))).))).(((((..........))))) ( -37.30) >DroVir_CAF1 794 120 + 1 UUCAUAUCGCAGCCGGCGCGUACCGCAAUGCGCUGCGUGGCGGAAUCGAACUGAAAGUAGCAGUUGGGCUGGCAGGCGUGCUCCAUCAUGCAGGCCUGCCGAAAGAGUCCGCUCAGCUCC ((((..(((...(((.((((((.(((...))).)))))).)))...)))..))))......((((((((.(((((((.(((........))).)))))))((.....)).)))))))).. ( -54.60) >DroGri_CAF1 809 120 + 1 UUCAGAUCGCAGCCGGCGCGCACAGCAAUCCGUUGCGUGGCCGCAUCGAAUUGAAAGUAGCAAUUGGGCUGGCAGGCGUGCUCCAUCAUGCAGGCCUGCCGAAAGACGCCGCUCAGCUCC (((((.(((..((.(((.(((.((((.....)))).))))))))..))).)))))..........((((((((((((.(((........))).))))))(....)........)))))). ( -49.00) >DroWil_CAF1 809 120 + 1 UUCAAAUUGCAACCGGCUCGCACUGCAAUCUGCUGGGUGGCCGCAUCGAAUGUGAAAUAACAAUUCGGUUGACAGGCAUGCUCCAUCAUGCAAGCCUGCCGGAAUAGGCCACUCAAUUCC ..((.((((((....(....)..)))))).)).(((((((((..(((((((((......)).)))))))((.(((((.(((........))).))))).)).....)))))))))..... ( -41.70) >DroMoj_CAF1 791 120 + 1 UUCAGGUCGCAGCCCGCACGCACCGCAAUGCGUUGUGUGGCUGCAUCGAACUGGAAGUAGCAAUUUGGCUGGCAGGCGUGCUCCAUCAUACAGGCUUGCCGGAAGAGACCGCUCAACUCU ....((((((((((((((((((......)))).)))).)))))).(((((.((.......)).)))))(((((((((.((..........)).)))))))))....)))).......... ( -48.20) >DroAna_CAF1 806 120 + 1 UGCAGGUCACACCCUGCCCGCACAGUGAUCUGCUGGGUGAAGGCGUCGAACUGGAAGUAGCAGUUGGGCUGGCAGGCGUGCUCCAUCAUGCAGGCCUGUCGGAACAGACCACUCAGCUCC .(((((((((....((....))..))))))))).((((..((..(((.(((((.......)))))(..(((((((((.(((........))).)))))))))..).)))..))..)))). ( -50.80) >consensus UUCAGAUCGCAGCCGGCACGCACAGCAAUCCGCUGCGUGGCGGCAUCGAACUGAAAGUAGCAAUUGGGCUGGCAGGCGUGCUCCAUCAUGCAGGCCUGCCGGAAGAGACCGCUCAGCUCC ...............((.......)).....((((.((((.....((.(((((.......))))).))(((((((((.(((........))).)))))))))......)))).))))... (-27.67 = -28.53 + 0.87)

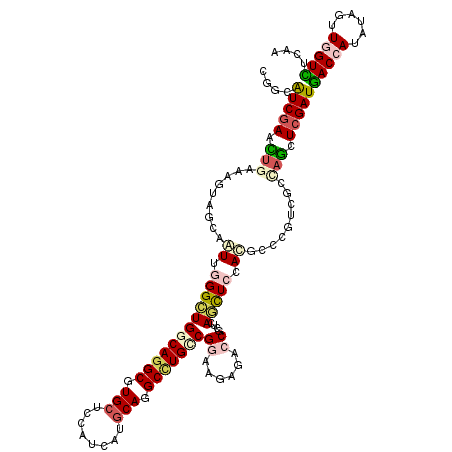
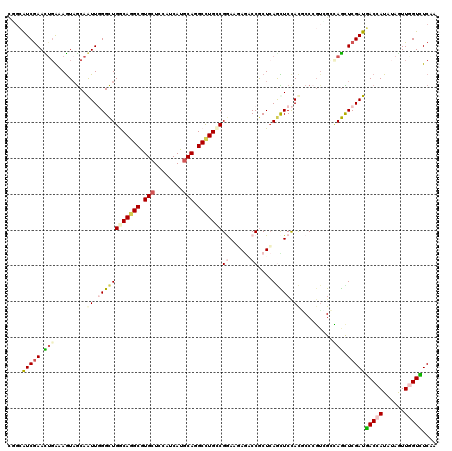
| Location | 1,587,082 – 1,587,202 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -27.72 |
| Energy contribution | -28.80 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1587082 120 + 20766785 UGUUAUCAAACUGAAAGUCGCAGUUGGGCUGACAAGCAUGCUCCAUCAUGCAGGCCUGCCGGAACAACCCGCUGAGUUCCACGCCACUCGGGAGUUCGAUAACCAUAAAGUUUGUUUCGA .((((((.((((..........((((..(((.((.((.(((........))).)).)).)))..))))...((((((........)))))).)))).))))))................. ( -31.40) >DroVir_CAF1 834 120 + 1 CGGAAUCGAACUGAAAGUAGCAGUUGGGCUGGCAGGCGUGCUCCAUCAUGCAGGCCUGCCGAAAGAGUCCGCUCAGCUCCACACCCGUCGCCAGCUCGAUGACCAUGUAGUUGGUCUCUA .((((((((.(((...(..((.(((((((((((((((.(((........))).)))))))....(((....)))))).))).))..))..)))).)))))(((((......)))))))). ( -49.50) >DroGri_CAF1 849 120 + 1 CCGCAUCGAAUUGAAAGUAGCAAUUGGGCUGGCAGGCGUGCUCCAUCAUGCAGGCCUGCCGAAAGACGCCGCUCAGCUCCACGCCCGUUGCUAGCUCGAUGACCAUGUAGUUAGUCUCAA ..((((.(.(((((...(((((((.((((((((((((.(((........))).))))))))...((.((......))))...)))))))))))..)))))..).))))............ ( -44.50) >DroWil_CAF1 849 120 + 1 CCGCAUCGAAUGUGAAAUAACAAUUCGGUUGACAGGCAUGCUCCAUCAUGCAAGCCUGCCGGAAUAGGCCACUCAAUUCCACUCCAGUGGGCAAUUCGAUGACCAUGUAAUUGGUCUCAA ....(((((((............(((((....(((((.(((........))).))))))))))...(.(((((............))))).).)))))))(((((......))))).... ( -37.90) >DroMoj_CAF1 831 120 + 1 CUGCAUCGAACUGGAAGUAGCAAUUUGGCUGGCAGGCGUGCUCCAUCAUACAGGCUUGCCGGAAGAGACCGCUCAACUCUAUGCCCGUGGCCAGCUCGAUGACCAUAUAGUUGGUCUCAA ....(((((.((((.....(((......(((((((((.((..........)).)))))))))..(((....))).......)))......)))).)))))(((((......))))).... ( -41.10) >DroAna_CAF1 846 120 + 1 AGGCGUCGAACUGGAAGUAGCAGUUGGGCUGGCAGGCGUGCUCCAUCAUGCAGGCCUGUCGGAACAGACCACUCAGCUCCACUCCCGAAGGCAGCUCGAUGACCAUAUAGUUGGUUUCAA .((((((....(((.(((.(.(((((((..(((((((.(((........))).)))))))((......)).)))))))).))).)))..))).))).((.(((((......))))))).. ( -44.30) >consensus CGGCAUCGAACUGAAAGUAGCAAUUGGGCUGGCAGGCGUGCUCCAUCAUGCAGGCCUGCCGGAAGAGACCGCUCAGCUCCACGCCCGUCGCCAGCUCGAUGACCAUAUAGUUGGUCUCAA ....(((((.(((.........((.((((((((((((.(((........))).)))))))((......))....))))).)).........))).)))))(((((......))))).... (-27.72 = -28.80 + 1.09)
| Location | 1,587,082 – 1,587,202 |
|---|---|
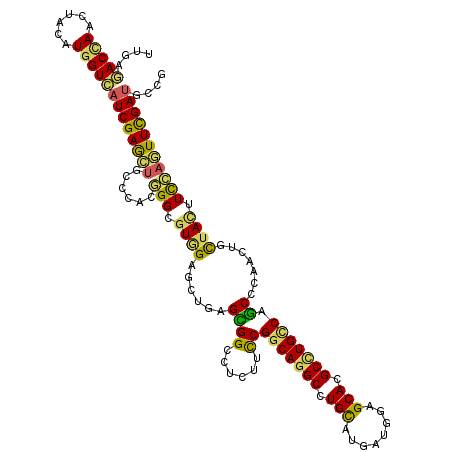
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -31.71 |
| Energy contribution | -30.64 |
| Covariance contribution | -1.07 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1587082 120 - 20766785 UCGAAACAAACUUUAUGGUUAUCGAACUCCCGAGUGGCGUGGAACUCAGCGGGUUGUUCCGGCAGGCCUGCAUGAUGGAGCAUGCUUGUCAGCCCAACUGCGACUUUCAGUUUGAUAACA .................(((((((((((...((((.((..(((((..........)))))(((.(((..(((((......)))))..))).))).....)).))))..))))))))))). ( -41.70) >DroVir_CAF1 834 120 - 1 UAGAGACCAACUACAUGGUCAUCGAGCUGGCGACGGGUGUGGAGCUGAGCGGACUCUUUCGGCAGGCCUGCAUGAUGGAGCACGCCUGCCAGCCCAACUGCUACUUUCAGUUCGAUUCCG ....(((((......)))))((((((((((.....(((.(((.((((((....)))....(((((((.(((........))).)))))))))))))...)))....)))))))))).... ( -49.60) >DroGri_CAF1 849 120 - 1 UUGAGACUAACUACAUGGUCAUCGAGCUAGCAACGGGCGUGGAGCUGAGCGGCGUCUUUCGGCAGGCCUGCAUGAUGGAGCACGCCUGCCAGCCCAAUUGCUACUUUCAAUUCGAUGCGG (((((((((......)))))...(((.((((((.(((((.((((((....))).)))..)(((((((.(((........))).))))))).))))..))))))))))))).......... ( -45.40) >DroWil_CAF1 849 120 - 1 UUGAGACCAAUUACAUGGUCAUCGAAUUGCCCACUGGAGUGGAAUUGAGUGGCCUAUUCCGGCAGGCUUGCAUGAUGGAGCAUGCCUGUCAACCGAAUUGUUAUUUCACAUUCGAUGCGG (((((((((......))))).))))..(((((((....)))).((((((((.........(((((((.(((........))).)))))))....(((.......))).))))))))))). ( -40.90) >DroMoj_CAF1 831 120 - 1 UUGAGACCAACUAUAUGGUCAUCGAGCUGGCCACGGGCAUAGAGUUGAGCGGUCUCUUCCGGCAAGCCUGUAUGAUGGAGCACGCCUGCCAGCCAAAUUGCUACUUCCAGUUCGAUGCAG ....(((((......)))))((((((((((.....((((.....(((.((((......))((((.((.(((........))).)).)))).)))))..))))....)))))))))).... ( -44.10) >DroAna_CAF1 846 120 - 1 UUGAAACCAACUAUAUGGUCAUCGAGCUGCCUUCGGGAGUGGAGCUGAGUGGUCUGUUCCGACAGGCCUGCAUGAUGGAGCACGCCUGCCAGCCCAACUGCUACUUCCAGUUCGACGCCU ................(((..((((((((((....))(((((.((((.(((((.(((((((.((.(....).)).))))))).))).)))))))).)))........)))))))).))). ( -44.00) >consensus UUGAGACCAACUACAUGGUCAUCGAGCUGCCCACGGGCGUGGAGCUGAGCGGCCUCUUCCGGCAGGCCUGCAUGAUGGAGCACGCCUGCCAGCCCAACUGCUACUUCCAGUUCGAUGCCG ....(((((......)))))((((((((......(((.((((......((((......))(((((((.(((........))).))))))).)).......)))).))))))))))).... (-31.71 = -30.64 + -1.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:10 2006