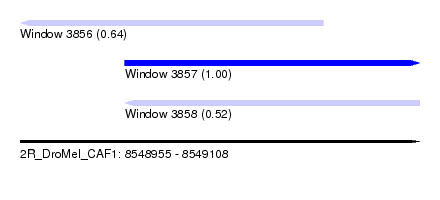
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,548,955 – 8,549,108 |
| Length | 153 |
| Max. P | 0.996023 |

| Location | 8,548,955 – 8,549,071 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.04 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.92 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8548955 116 - 20766785 ACAAACAAAUACGACGCACACACACUUUUGUGCAUUUAUAUUUGUUCAUAA----UCGUAAUUAUUUCAGGCCAAGCAGUUGGACGCAUUGAUGUAAUGAUAAACAAAGGAAUCAUUUCA ...((((((((..(.(((((........)))))..)..))))))))....(----((((...(((.((((.((((....)))).....)))).))))))))........(((....))). ( -18.60) >DroSec_CAF1 24480 120 - 1 ACAAACAAAUACGACGCACACACACGUUAGUGCAUUUAUAUUUGUUCAUAUUUGUUCGUAAUUAUUUCAGGCCAAGCAGUUGGACGCAUUGAAGUAAUGAUAAACAAAGGAAUCAUUUCA ...((((((((..(.((((..........))))..)..))))))))....((((((..(.((((((((((.((((....)))).....)))))))))).)..)))))).(((....))). ( -25.50) >DroSim_CAF1 28119 116 - 1 ACAAACAAAUACGACGCACACACACGUUAGUGCAUUUAUAUUUGUUCAUAA----UCGUAAUUAUUUCAGGCCAAGCAGUUGGACGCAUUGAAGUAAUGAUAAACAAAGGAAUCAUUUCA ...((((((((..(.((((..........))))..)..))))))))....(----((((...((((((((.((((....)))).....)))))))))))))........(((....))). ( -20.10) >DroEre_CAF1 13651 114 - 1 ACAAACAAAUACGACGCACACA--CUUAUGUGCAUUUAUAUUUGUUCAUAA----UCGUAAUUAUUUCAGGCCAAGCAGUUGGACGCAUUGAAGUAAUGAUAAACAAAGGAAUCAUUUCA ...((((((((..(.(((((..--....)))))..)..))))))))....(----((((...((((((((.((((....)))).....)))))))))))))........(((....))). ( -21.90) >DroYak_CAF1 27759 114 - 1 ACAAACAAAUACGAGGCACACA--GUUAUGUGCAUUUAUAUUUGUUCAUAA----UCGUAAUUAUUUCAGGCCCAGCAGUUGGACGCACUGAAGUAAUGAUAAACAAAGGAAUCAUUUCA ...((((((((..(((((((..--....))))).))..))))))))....(----((((...((((((((((((((...))))..)).)))))))))))))........(((....))). ( -26.80) >consensus ACAAACAAAUACGACGCACACACACUUUUGUGCAUUUAUAUUUGUUCAUAA____UCGUAAUUAUUUCAGGCCAAGCAGUUGGACGCAUUGAAGUAAUGAUAAACAAAGGAAUCAUUUCA ...((((((((..(.(((((........)))))..)..))))))))..............((((((((((.((((....)))).....))))))))))...........(((....))). (-19.28 = -19.92 + 0.64)

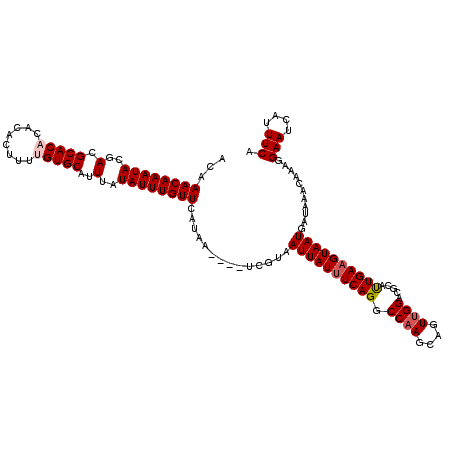
| Location | 8,548,995 – 8,549,108 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.21 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -25.22 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8548995 113 + 20766785 ACUGCUUGGCCUGAAAUAAUUACGA----UUAUGAACAAAUAUAAAUGCACAAAAGUGUGUGUGCGUCGUAUUUGUUUGUAAAUACA--CA-AUGCUGGCGAAUGAAGGCCUUUGAAGAU .((....(((((......(((.((.----(((..(((((((((..(((((((........))))))).)))))))))..))).....--((-....)).)))))..))))).....)).. ( -28.60) >DroSec_CAF1 24520 117 + 1 ACUGCUUGGCCUGAAAUAAUUACGAACAAAUAUGAACAAAUAUAAAUGCACUAACGUGUGUGUGCGUCGUAUUUGUUUGUAAAUACA--CA-AAGCUGGCGAAUGAAGGCCUUUGAAGAU .((....(((((.......((((((((((((((((....((((....((((....))))))))...)))))))))))))))).....--((-....))........))))).....)).. ( -32.90) >DroSim_CAF1 28159 114 + 1 ACUGCUUGGCCUGAAAUAAUUACGA----UUAUGAACAAAUAUAAAUGCACUAACGUGUGUGUGCGUCGUAUUUGUUUGUAAAUACA--AAAAAGCUGGCGAAUGAAGGCCUUUGAAGAU .((....(((((......(((.((.----(((..(((((((((..((((((..........)))))).)))))))))..)))...((--.......)).)))))..))))).....)).. ( -27.00) >DroEre_CAF1 13691 111 + 1 ACUGCUUGGCCUGAAAUAAUUACGA----UUAUGAACAAAUAUAAAUGCACAUAAG--UGUGUGCGUCGUAUUUGUUUGUAAAUACA--CA-ACGCUGGCGAAUGAAGGCCUUUGAAGAU .((....(((((.............----(((..(((((((((..((((((((...--.)))))))).)))))))))..))).....--..-.((....)).....))))).....)).. ( -31.90) >DroYak_CAF1 27799 113 + 1 ACUGCUGGGCCUGAAAUAAUUACGA----UUAUGAACAAAUAUAAAUGCACAUAAC--UGUGUGCCUCGUAUUUGUUUGUAAAUACACACA-GCGCUGGCGAAUGAAGGCCUUUGAAGAU .((...((((((.............----(((..(((((((((....((((((...--.))))))...)))))))))..))).........-((....))......))))))....)).. ( -31.60) >consensus ACUGCUUGGCCUGAAAUAAUUACGA____UUAUGAACAAAUAUAAAUGCACAAAAGUGUGUGUGCGUCGUAUUUGUUUGUAAAUACA__CA_AAGCUGGCGAAUGAAGGCCUUUGAAGAU .((....(((((.................(((..(((((((((..(((((((........))))))).)))))))))..)))............(....)......))))).....)).. (-25.22 = -26.02 + 0.80)

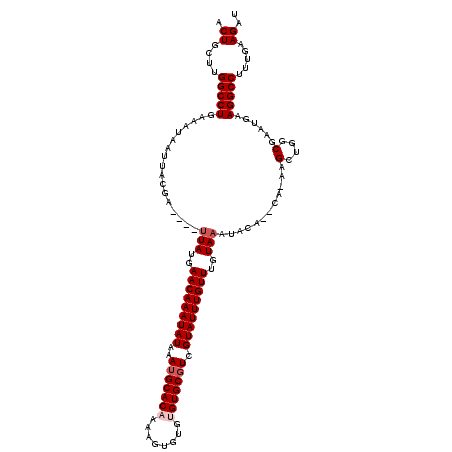
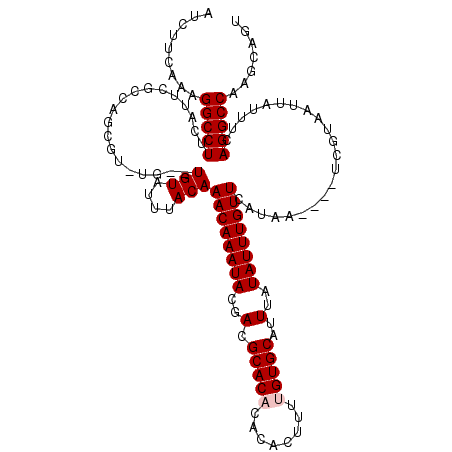
| Location | 8,548,995 – 8,549,108 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.21 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.34 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8548995 113 - 20766785 AUCUUCAAAGGCCUUCAUUCGCCAGCAU-UG--UGUAUUUACAAACAAAUACGACGCACACACACUUUUGUGCAUUUAUAUUUGUUCAUAA----UCGUAAUUAUUUCAGGCCAAGCAGU .........(((((..........((((-((--((........((((((((..(.(((((........)))))..)..)))))))))))))----).)).........)))))....... ( -22.11) >DroSec_CAF1 24520 117 - 1 AUCUUCAAAGGCCUUCAUUCGCCAGCUU-UG--UGUAUUUACAAACAAAUACGACGCACACACACGUUAGUGCAUUUAUAUUUGUUCAUAUUUGUUCGUAAUUAUUUCAGGCCAAGCAGU .........(((........))).((((-.(--((((.((((.((((((((.((((........)))).(.(((........))).).)))))))).)))).))).....)).))))... ( -21.30) >DroSim_CAF1 28159 114 - 1 AUCUUCAAAGGCCUUCAUUCGCCAGCUUUUU--UGUAUUUACAAACAAAUACGACGCACACACACGUUAGUGCAUUUAUAUUUGUUCAUAA----UCGUAAUUAUUUCAGGCCAAGCAGU .........(((((..........((.....--.))..(((..((((((((..(.((((..........))))..)..))))))))..)))----.............)))))....... ( -18.70) >DroEre_CAF1 13691 111 - 1 AUCUUCAAAGGCCUUCAUUCGCCAGCGU-UG--UGUAUUUACAAACAAAUACGACGCACACA--CUUAUGUGCAUUUAUAUUUGUUCAUAA----UCGUAAUUAUUUCAGGCCAAGCAGU .........(((((..........((((-((--(((............)))))))))..((.--.(((((.(((........))).)))))----..)).........)))))....... ( -25.30) >DroYak_CAF1 27799 113 - 1 AUCUUCAAAGGCCUUCAUUCGCCAGCGC-UGUGUGUAUUUACAAACAAAUACGAGGCACACA--GUUAUGUGCAUUUAUAUUUGUUCAUAA----UCGUAAUUAUUUCAGGCCCAGCAGU .........(((((..........(.((-(...(((((((......))))))).))).)((.--((((((.(((........))).)))))----).)).........)))))....... ( -24.40) >consensus AUCUUCAAAGGCCUUCAUUCGCCAGCGU_UG__UGUAUUUACAAACAAAUACGACGCACACACACUUUUGUGCAUUUAUAUUUGUUCAUAA____UCGUAAUUAUUUCAGGCCAAGCAGU .........(((((...................(((....)))((((((((..(.(((((........)))))..)..))))))))......................)))))....... (-17.94 = -18.34 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:21 2006