
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
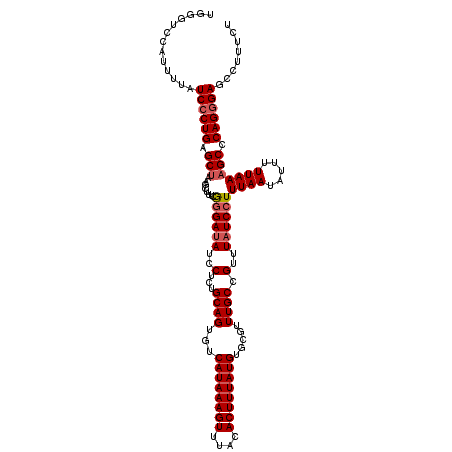
| Location | 8,545,326 – 8,545,440 |
| Length | 114 |
| Max. P | 0.999987 |

| Location | 8,545,326 – 8,545,440 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -26.06 |
| Energy contribution | -26.70 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
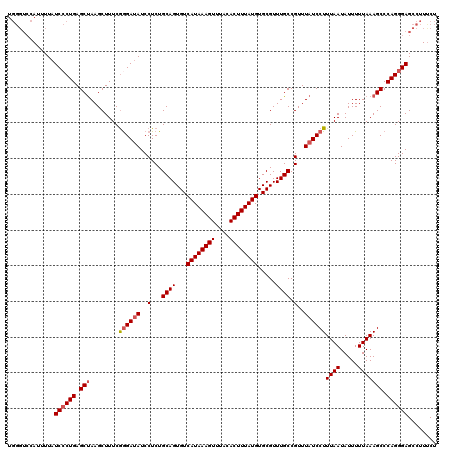
>2R_DroMel_CAF1 8545326 114 + 20766785 ------CAUUUUAUCACUGAGCUAAGCUUUCGGGAUAUCCUCUGCAGUGUCAUAAAGUUUACACUUUAUGUGCGUUUGCCGUUUAUCCUUUAAUAUUUUUAAAGCCCAGGGAGCCUUUCU ------...................(((..((((.........((((...((((((((....)))))))).....))))........((((((.....))))))))).)..)))...... ( -23.60) >DroSec_CAF1 20947 120 + 1 UGGGUCCAUGUUAUCCCUGAGCUAAGCUUUCAAGAUAUCCUCUGCAGUGUCAUAAAGUUUACACUUUAUGUGCGUUUGCCGUUUAUCCUUUAAUAUUUUUAAAGCCCAGGGAGCCUUUCU .((((........((((((.(((..........((((..(...((((...((((((((....)))))))).....)))).)..))))..((((.....))))))).)))))))))).... ( -27.30) >DroSim_CAF1 24558 120 + 1 UGGGUCCAUGUUAUCCCUGAGCCAAGCUUUCGGGAUAUCCUCUGCAGUGUCAUAAAGUUUACACUUUAUGUGCGUUUGCCGUUUAUCCUUUAAUAUUUUUAAAGCCCAGGGAGCCUUUCU .((.(((....((((((.((((...))))..))))))....(((..(((.((((((((....)))))))))))..............((((((.....))))))..)))))).))..... ( -30.50) >DroEre_CAF1 10251 120 + 1 UGGGUCCAUUUUAUCCCUGAGCUAAGCUUUCGGGAAAUCCUCUGCAGUGUCAUAAAGUUUACACUUUAUGUGCGUUUGCCGUUUAUCCUUUAAUAUUUUUAAAGCCCAGGGAGCCUUCCU .((((........((((((.(((((((....(((....)))..((((...((((((((....)))))))).....)))).)))))...(((((.....))))))).)))))))))).... ( -28.60) >DroYak_CAF1 24239 120 + 1 UGGGUCCAUUUUAUCCCUGAGCUAAGCUUUCGGGAUAUCCUCUGCAGUGUCAUAAAGUUUACACUUUAUGUGCGUUUGCCGUUUAUCCUUUAAUAUUUUUAAAGCCCAGGGAGCCUUUCU .((((........((((((.(((........((((((..(...((((...((((((((....)))))))).....)))).)..))))))((((.....))))))).)))))))))).... ( -31.60) >consensus UGGGUCCAUUUUAUCCCUGAGCUAAGCUUUCGGGAUAUCCUCUGCAGUGUCAUAAAGUUUACACUUUAUGUGCGUUUGCCGUUUAUCCUUUAAUAUUUUUAAAGCCCAGGGAGCCUUUCU .............((((((.(((........((((((..(...((((...((((((((....)))))))).....)))).)..))))))((((.....))))))).))))))........ (-26.06 = -26.70 + 0.64)

| Location | 8,545,326 – 8,545,440 |
|---|---|
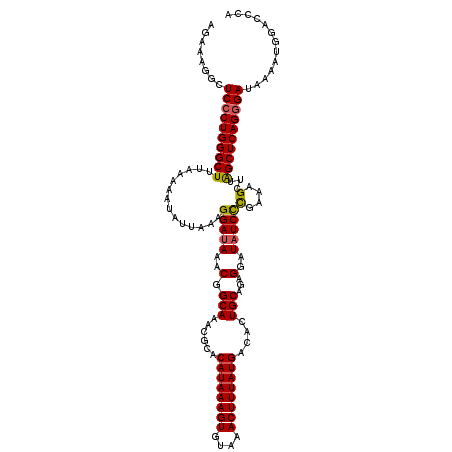
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -31.44 |
| Energy contribution | -31.36 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.82 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.44 |
| SVM RNA-class probability | 0.999987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
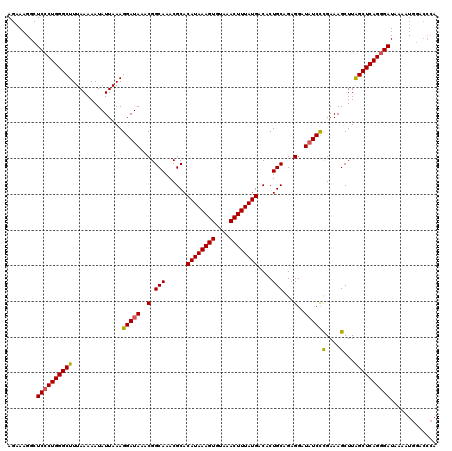
>2R_DroMel_CAF1 8545326 114 - 20766785 AGAAAGGCUCCCUGGGCUUUAAAAAUAUUAAAGGAUAAACGGCAAACGCACAUAAAGUGUAAACUUUAUGACACUGCAGAGGAUAUCCCGAAAGCUUAGCUCAGUGAUAAAAUG------ ........((.(((((((..............(((((..(.(((......((((((((....))))))))....)))...)..)))))(....)...))))))).)).......------ ( -27.10) >DroSec_CAF1 20947 120 - 1 AGAAAGGCUCCCUGGGCUUUAAAAAUAUUAAAGGAUAAACGGCAAACGCACAUAAAGUGUAAACUUUAUGACACUGCAGAGGAUAUCUUGAAAGCUUAGCUCAGGGAUAACAUGGACCCA ........((((((((((.............((((((..(.(((......((((((((....))))))))....)))...)..))))))........))))))))))............. ( -31.70) >DroSim_CAF1 24558 120 - 1 AGAAAGGCUCCCUGGGCUUUAAAAAUAUUAAAGGAUAAACGGCAAACGCACAUAAAGUGUAAACUUUAUGACACUGCAGAGGAUAUCCCGAAAGCUUGGCUCAGGGAUAACAUGGACCCA ........((((((((((..............(((((..(.(((......((((((((....))))))))....)))...)..)))))(....)...))))))))))............. ( -34.20) >DroEre_CAF1 10251 120 - 1 AGGAAGGCUCCCUGGGCUUUAAAAAUAUUAAAGGAUAAACGGCAAACGCACAUAAAGUGUAAACUUUAUGACACUGCAGAGGAUUUCCCGAAAGCUUAGCUCAGGGAUAAAAUGGACCCA ........((((((((((.......((((....))))...(((....(((((((((((....))))))))....))).(.((....)))....))).))))))))))............. ( -30.70) >DroYak_CAF1 24239 120 - 1 AGAAAGGCUCCCUGGGCUUUAAAAAUAUUAAAGGAUAAACGGCAAACGCACAUAAAGUGUAAACUUUAUGACACUGCAGAGGAUAUCCCGAAAGCUUAGCUCAGGGAUAAAAUGGACCCA ........((((((((((..............(((((..(.(((......((((((((....))))))))....)))...)..)))))(....)...))))))))))............. ( -34.20) >consensus AGAAAGGCUCCCUGGGCUUUAAAAAUAUUAAAGGAUAAACGGCAAACGCACAUAAAGUGUAAACUUUAUGACACUGCAGAGGAUAUCCCGAAAGCUUAGCUCAGGGAUAAAAUGGACCCA ........((((((((((..............(((((..(.(((......((((((((....))))))))....)))...)..)))))(....)...))))))))))............. (-31.44 = -31.36 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:15 2006