| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,542,700 – 8,542,894 |
| Length | 194 |
| Max. P | 0.677955 |

| Location | 8,542,700 – 8,542,815 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.81 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -27.45 |
| Energy contribution | -28.05 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8542700 115 - 20766785 CCACCG---CUUCGAAUGGGUUUUCUUUUCUAUUCGCUUGAGCGGGCAC-UGGCCAAGAAAAAUGUUGGGUUAGCGUGGUUAUCCGCCGCGU-UUUGGCCACAUUGCUUUAAGUCGGUGA ...(((---(((((((((((........))))))))...)))))).(((-((((.(((...(((((.((.(.((((((((.....)))))))-)..).))))))).)))...))))))). ( -44.90) >DroSec_CAF1 18352 110 - 1 -CACCG---CUCAGAAUGGGUUUUCUUUUCUAUUCGCUUCAGUGGGCAC-UGGCCAAGAAAAAUUUUGGGUUAGCGUGGUUAUCCGCCGCGU-UUUGGCCACAUUGCUUUAAGUCA---- -..(((---((..(((((((........))))))).....)))))(((.-((((((((...............(((((((.....)))))))-))))))))...))).........---- ( -33.36) >DroSim_CAF1 21955 110 - 1 -CACCG---CUCAGAAUGGGUUUUCUUUUCUAUUCGCUUCAGUGGGCAC-UGGCCAAGAAAAAUGUUGGGUUAGCGUGGUUAUCCGCCGCGU-UUUGGCCACAUUGCUUUAAGUCA---- -..(((---((..(((((((........))))))).....)))))(((.-(((((((((.....((((...))))(((((.....))))).)-))))))))...))).........---- ( -33.60) >DroEre_CAF1 7650 107 - 1 -GAAUGAGUAUGAGAAUGGGUUUU-----CUAUUCGCUUGAGCGGUCAC-UGGCCAAGAAAA--GUUGGGUUAGCGUGGUUAUCCGCCGCCUUUUUGGCCACAUUGCUUUAAGUCA---- -....(((((.(((((....))))-----))))))(((((((((((...-((((((((((..--((((...))))(((((.....))))).)))))))))).))))).))))))..---- ( -33.30) >DroYak_CAF1 21520 100 - 1 -AA---------AGAAUGGGUUUUC---UCUAUUCGCUUCAGCGGGCACAUGGCCAAGAAAC--GUUGGGUUAGCGUGGUUAUCCGCCGCGU-UUUGGCCACAUUGCUUUAAGUCG---- -..---------.(((((((.....---)))))))((....))(((((..(((((((((..(--(.(((((.(((...)))))))).))..)-))))))))...))))).......---- ( -31.60) >consensus _CACCG___CUCAGAAUGGGUUUUCUUUUCUAUUCGCUUCAGCGGGCAC_UGGCCAAGAAAAAUGUUGGGUUAGCGUGGUUAUCCGCCGCGU_UUUGGCCACAUUGCUUUAAGUCA____ .............(((((((........)))))))((((....(((((..((((((((...............(((((((.....))))))).))))))))...))))).))))...... (-27.45 = -28.05 + 0.60)

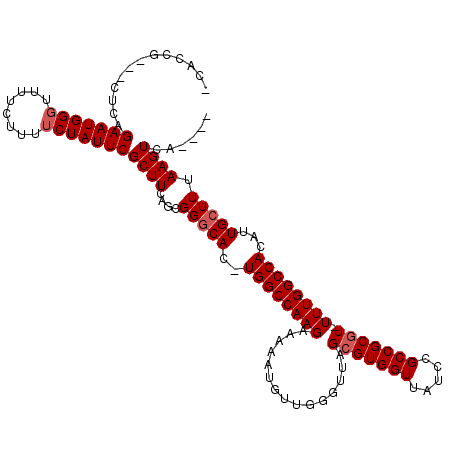
| Location | 8,542,778 – 8,542,894 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.44 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -19.20 |
| Energy contribution | -20.00 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8542778 116 - 20766785 UGGUGUGCUUGGUGGUGCAAUUAACCGCAAAAGGUUUCCUUUUGUCACUCACUGGAUCUACCGGGCCUCUC-CUCGGUCACCACCG---CUUCGAAUGGGUUUUCUUUUCUAUUCGCUUG ......((..(((((((.........((((((((...))))))))..............((((((......-))))))))))))).---....(((((((........)))))))))... ( -34.30) >DroSec_CAF1 18426 111 - 1 UGGUGUGGUUGGUGGUGCAAUUAACCGCAAAAGGUUUCCUUUUGUCACUCGCUGGGUCU-CCGGGCCUCUCGCUCG-----CACCG---CUCAGAAUGGGUUUUCUUUUCUAUUCGCUUC .(((.....(((((((((........((((((((...)))))))).....((.(((((.-...)))))...))..)-----)))))---).))(((((((........)))))))))).. ( -35.10) >DroSim_CAF1 22029 111 - 1 UGGUGUGGUUGGUGGUGCAAUUAACCGCAAAAGGUUUCCUUUUGUCACUCGCUGGGUCU-CCGGGCCUCUCGCUCG-----CACCG---CUCAGAAUGGGUUUUCUUUUCUAUUCGCUUC .(((.....(((((((((........((((((((...)))))))).....((.(((((.-...)))))...))..)-----)))))---).))(((((((........)))))))))).. ( -35.10) >DroEre_CAF1 7723 100 - 1 UAGUC-GG-CGGUGGUGCAAUUAACCACAAAAGGUUUCCUUUUGUCACUCACUGGGC--------CCUCUCGCUCA-----GAAUGAGUAUGAGAAUGGGUUUU-----CUAUUCGCUUG .....-((-(.(((((.......)))))((((((...)))))))))(((((((((((--------......)))))-----)..)))))....((((((.....-----))))))..... ( -30.70) >DroYak_CAF1 21593 91 - 1 CAGUC-AGUCGGUGGUGCAAUUAACCACAAAAGGUUUCCUUUUGUCACUCAUUCG-----------CUGGCACUCG-----AA---------AGAAUGGGUUUUC---UCUAUUCGCUUC ..(((-(((..(((((((((..((((......)))).....))).))).)))..)-----------)))))....(-----((---------.(((((((.....---)))))))..))) ( -26.20) >consensus UGGUGUGGUUGGUGGUGCAAUUAACCGCAAAAGGUUUCCUUUUGUCACUCACUGGGUCU_CCGGGCCUCUCGCUCG_____CACCG___CUCAGAAUGGGUUUUCUUUUCUAUUCGCUUC .((((....(((((((((((..((((......)))).....))).))).)))))((((.....))))..............))))........(((((((........)))))))..... (-19.20 = -20.00 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:11 2006