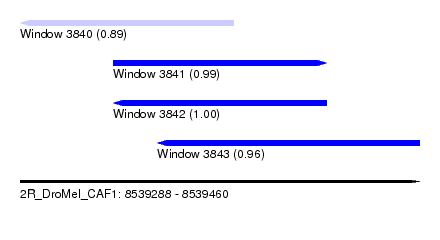
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,539,288 – 8,539,460 |
| Length | 172 |
| Max. P | 0.997684 |

| Location | 8,539,288 – 8,539,380 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.03 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -25.34 |
| Energy contribution | -26.54 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8539288 92 - 20766785 AAUCGCGAUACCAAACGAGCGGCGAGAGCGACAAG---------------------CGGCAAGCGGCAAGAUACGAGAUUCGGUGCCGAAAGCCAGUUGCCAAUUGCGACGGC ..(((((((..(....)...(((((..((.....)---------------------)(((...(((((.((........))..)))))...)))..))))).))))))).... ( -30.40) >DroSec_CAF1 14865 85 - 1 AAUCGCGAUACCAAACGAGUGGCGAGAGC----------------------------GGCGAGCGGCAAGAUUCGAGAUUCGGUGCCGAAAGCCAGUUGCCAAUUGCGACGGC ..(((((((....(((.....((....))----------------------------(((...(((((.((........))..)))))...))).)))....))))))).... ( -30.60) >DroSim_CAF1 18391 92 - 1 AAUCGCGAUACCAAACGAGUGGCGAGAGCGGCAAG---------------------CGGCGAGCGGCAAGAUUCGAGAUUCGGUGCCGAAAGCCAGUUGCCAAUUGCGACGGC ..(((((((............((....))(((((.---------------------.(((...(((((.((........))..)))))...)))..))))).))))))).... ( -36.70) >DroEre_CAF1 4158 98 - 1 AAUCGCGAUACCA-ACGAGCGGCAAGAGCAGCGAGC--------------UGCAAGCCGCAAGCGGCAAGAUUCGAGAUUCGGUGCCGAAAGCCAGUUGCCAAUUGCGACGGC ..(((((((..((-((..(((((....((((....)--------------)))..)))))...(((((.((........))..))))).......))))...))))))).... ( -36.80) >DroYak_CAF1 18080 112 - 1 AAUCGCGAUACCA-GCGAGCGGCGAGAGCAGCAAGCAGCAAGUUGCAAGUUGCAAGCCGCAAGUCGCAAGAUUCAAGAUUCGGUGCCGAAAGCCAGUUGCCAAUUGCGACGGC ..((((.......-))))(((((....(((((..((((....))))..)))))..))))).((((....)))).....((((....)))).(((.(((((.....)))))))) ( -43.40) >consensus AAUCGCGAUACCAAACGAGCGGCGAGAGCAGCAAG_____________________CGGCAAGCGGCAAGAUUCGAGAUUCGGUGCCGAAAGCCAGUUGCCAAUUGCGACGGC ..(((((((....(((..((.((....)).)).........................(((...(((((.((........))..)))))...))).)))....))))))).... (-25.34 = -26.54 + 1.20)

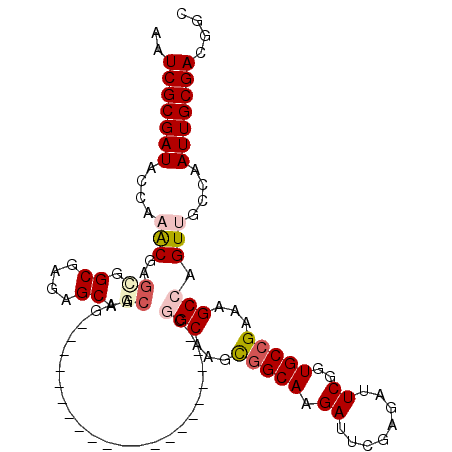
| Location | 8,539,328 – 8,539,420 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -20.38 |
| Energy contribution | -20.18 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8539328 92 + 20766785 UAUCUUGCCGCUUGCCG---------------------CUUGUCGCUCUCGCCGCUCGUUUGGUAUCGCGAUUGAAAUCGCAAUUAAGUUUUAUUUUGUGGCGAGCAUUUUUU .........((((((((---------------------(.((..(((...((((......))))...(((((....))))).....)))..))....)))))))))....... ( -25.90) >DroSec_CAF1 14905 85 + 1 AAUCUUGCCGCUCGCC----------------------------GCUCUCGCCACUCGUUUGGUAUCGCGAUUGAAAUCGCAAUUAAGUUUUAUUUUGUGGCGAGCAUUUUUU .........(((((((----------------------------((....((((......))))...(((((....)))))................)))))))))....... ( -28.70) >DroSim_CAF1 18431 92 + 1 AAUCUUGCCGCUCGCCG---------------------CUUGCCGCUCUCGCCACUCGUUUGGUAUCGCGAUUGAAAUCGCAAUUAAGUUUUAUUUUGUGGCGAGCAUUUUUU .........((((((((---------------------(...........((((......))))...(((((....)))))................)))))))))....... ( -28.70) >DroEre_CAF1 4198 98 + 1 AAUCUUGCCGCUUGCGGCUUGCA--------------GCUCGCUGCUCUUGCCGCUCGU-UGGUAUCGCGAUUGAAAUCGCAAUUAAGUUUUAUUUUGUGGCGAGCAUUUUUU ...((((((((..(((((..(((--------------(....))))....)))))....-.......(((((....)))))................))))))))........ ( -35.00) >DroYak_CAF1 18120 112 + 1 AAUCUUGCGACUUGCGGCUUGCAACUUGCAACUUGCUGCUUGCUGCUCUCGCCGCUCGC-UGGUAUCGCGAUUGAAAUCGCAAUUAAGUUUUAUUUUGUGGCGAGCAUUUUUU ......((((...(((((..(((....((.....)))))..)))))..)))).((((((-(......(((((....)))))...((((......)))).)))))))....... ( -36.50) >consensus AAUCUUGCCGCUUGCCG_____________________CUUGCCGCUCUCGCCGCUCGUUUGGUAUCGCGAUUGAAAUCGCAAUUAAGUUUUAUUUUGUGGCGAGCAUUUUUU ...((((((((.......................................((((......))))...(((((....)))))................))))))))........ (-20.38 = -20.18 + -0.20)

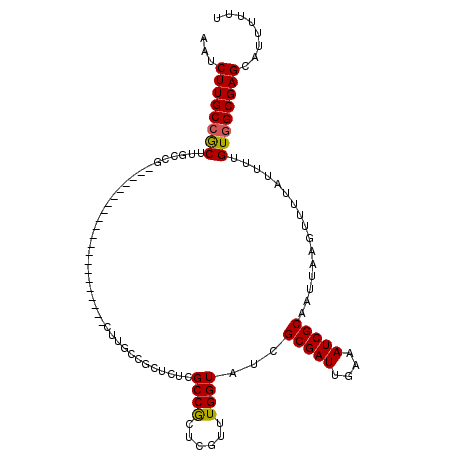
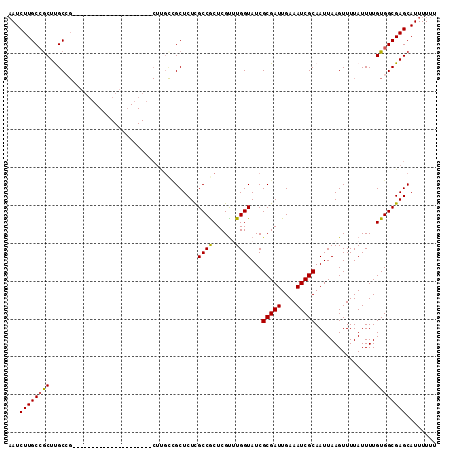
| Location | 8,539,328 – 8,539,420 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.98 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8539328 92 - 20766785 AAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCAAACGAGCGGCGAGAGCGACAAG---------------------CGGCAAGCGGCAAGAUA ......((((.(((...............((((((((....)))))))).........((.((....)).....)---------------------)))).))))........ ( -23.20) >DroSec_CAF1 14905 85 - 1 AAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCAAACGAGUGGCGAGAGC----------------------------GGCGAGCGGCAAGAUU ......((((((((...............((((((((....))))))))............((....))----------------------------))))))))........ ( -28.40) >DroSim_CAF1 18431 92 - 1 AAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCAAACGAGUGGCGAGAGCGGCAAG---------------------CGGCGAGCGGCAAGAUU ......((((((((.(.............((((((((....)))))))).........((.((....)).))..)---------------------.))))))))........ ( -32.40) >DroEre_CAF1 4198 98 - 1 AAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCA-ACGAGCGGCAAGAGCAGCGAGC--------------UGCAAGCCGCAAGCGGCAAGAUU ......(((.(((................((((((((....))))))))....-....(((((....((((....)--------------)))..)))))..))))))..... ( -31.90) >DroYak_CAF1 18120 112 - 1 AAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCA-GCGAGCGGCGAGAGCAGCAAGCAGCAAGUUGCAAGUUGCAAGCCGCAAGUCGCAAGAUU ..............................(((((((((...((((.......-))))(((((....(((((..((((....))))..)))))..)))))))))))))).... ( -36.90) >consensus AAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCAAACGAGCGGCGAGAGCAGCAAG_____________________CGGCAAGCGGCAAGAUU .......(((.(((...............((((((((....)))))))).........((.((....)).)).........................))).)))......... (-19.14 = -19.90 + 0.76)

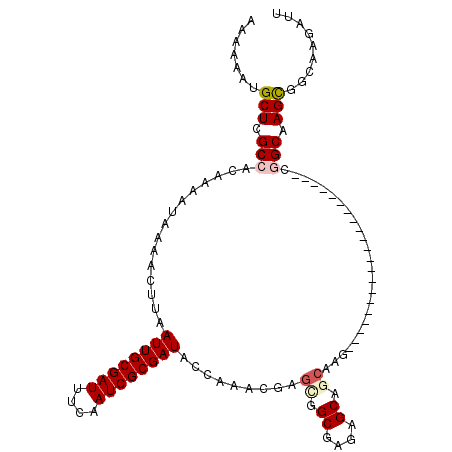
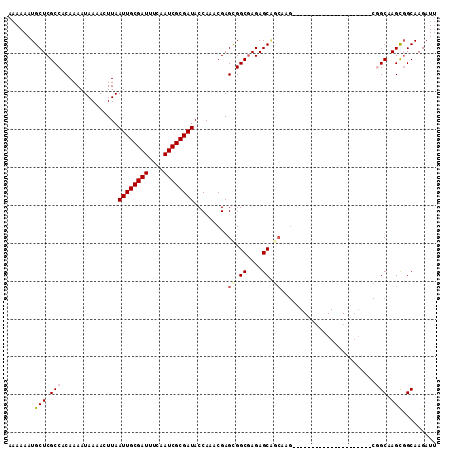
| Location | 8,539,347 – 8,539,460 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.57 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -28.04 |
| Energy contribution | -28.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8539347 113 - 20766785 AGCCGUUGCAAAUGGGGCAGUCGUCGCGUCGUUGCUAAUUAAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCAAACGAGCGGCGAGAGCGACA .(((.(.......).)))....(((((.((((((((..........((.......))............((((((((....))))))))........))))))))..))))). ( -32.20) >DroSec_CAF1 14921 109 - 1 AGCCGUUGCAAAUGGGGCAGUCGUCGCGUCGUUGCUAAUUAAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCAAACGAGUGGCGAGAGC---- ..((((.....))))(((((.((......)))))))............((((((((.............((((((((....))))))))..(....).))))))))...---- ( -30.40) >DroSim_CAF1 18450 113 - 1 AGCCGUUUCAAAUGGGGCAGUCGUCGCGUCGUUGCUAAUUAAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCAAACGAGUGGCGAGAGCGGCA .((((((........(((((.((......)))))))............((((((((.............((((((((....))))))))..(....).)))))))))))))). ( -37.60) >DroEre_CAF1 4224 112 - 1 AGCCGUUGCAAAUGGGGCAGUCGUCGCGUCGUUGCUAAUUAAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCA-ACGAGCGGCAAGAGCAGCG ...((((((.....((....))(((((.((((((............((.......))............((((((((....))))))))..))-)))))))))....)))))) ( -28.90) >DroYak_CAF1 18160 112 - 1 AGCCGUUGCAAAUGGGGCAGUCGUCGCGUCGUUGCUAAUUAAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCA-GCGAGCGGCGAGAGCAGCA ....(((((......(((((.((......)))))))............((((((...............((((((((....))))))))....-(....))))))).))))). ( -33.20) >consensus AGCCGUUGCAAAUGGGGCAGUCGUCGCGUCGUUGCUAAUUAAAAAAUGCUCGCCACAAAAUAAAACUUAAUUGCGAUUUCAAUCGCGAUACCAAACGAGCGGCGAGAGCAGCA ..((((.....)))).((..(((((((.(((((((............))....................((((((((....))))))))....))))))))))))..)).... (-28.04 = -28.24 + 0.20)

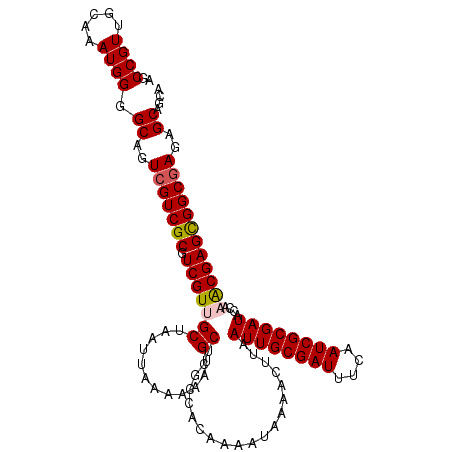
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:06 2006