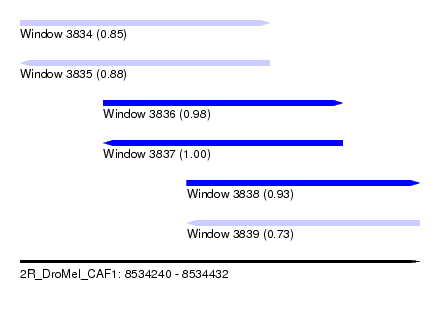
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,534,240 – 8,534,432 |
| Length | 192 |
| Max. P | 0.996366 |

| Location | 8,534,240 – 8,534,360 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.24 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -22.64 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8534240 120 + 20766785 AUGAAUAAACCAGGCACAACAACGCAUUUGAUUGAGUGUGUGUGUGUGUGCUCUGCUCAUUACAUUCAUUAACUUUAAUGAAUUGAUAUAUUCAGGCAAACUCGAAGGCCAUCAAAGGCC .((((((...(((((((((((((((((((....)))))))).))).))))).)))...((((.((((((((....)))))))))))).))))))............((((......)))) ( -35.50) >DroSec_CAF1 9746 111 + 1 AUGAAUAAACCAGGCACA---CCGCUUAUGAUUGAGUGUGU----GUGCGCUCUGCUCAUUACAUUCAUUAACUUUAAUGAAUUGAU--AUUCAGGCAAACUCGAAGGCCAACUGAGGCC .(((.........(((((---(((((((....)))))).))----))))((.(((...((((.((((((((....))))))))))))--...)))))....)))..((((......)))) ( -34.90) >DroSim_CAF1 13210 113 + 1 AUGAAUAAGCCAGGCACA---CCGCUUUUGAUUGAGUGUGUG--UGUGUGCUCUGCUCAUUACAUUCAUUAACUUUAAUGAAUUGAU--AUUCAGGCAAACUCGAAGGCCAAAUGAGGCC .(((((((((((.(((((---(.((((......)))))))))--).)).)))....(((....((((((((....))))))))))))--)))))............((((......)))) ( -32.70) >DroYak_CAF1 12983 105 + 1 AUGAAUAAACCAGGCA-----CCGCUUGUGAUUGAGUGU--------GUGCUCUGCUCAUUACAUUCAUUAAGUUUAAUGAAUUGAU--AUUCAGGCAAACUCGAAGGCCAUCCGAGGCC .((((((...((((((-----((((((......))))).--------)))).))).(((....((((((((....))))))))))))--)))))............((((......)))) ( -29.50) >consensus AUGAAUAAACCAGGCACA___CCGCUUUUGAUUGAGUGUGU____GUGUGCUCUGCUCAUUACAUUCAUUAACUUUAAUGAAUUGAU__AUUCAGGCAAACUCGAAGGCCAACUGAGGCC ............(((........((((((((..(((((..(....)..)))))((((......((((((((....))))))))(((.....)))))))...))))))))........))) (-22.64 = -23.27 + 0.63)

| Location | 8,534,240 – 8,534,360 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.24 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -24.15 |
| Energy contribution | -24.09 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8534240 120 - 20766785 GGCCUUUGAUGGCCUUCGAGUUUGCCUGAAUAUAUCAAUUCAUUAAAGUUAAUGAAUGUAAUGAGCAGAGCACACACACACACACUCAAUCAAAUGCGUUGUUGUGCCUGGUUUAUUCAU ((((......))))...((((..(((...........((((((((....)))))))).........((.(((((.(((.((.((..........)).)))))))))))))))..)))).. ( -29.20) >DroSec_CAF1 9746 111 - 1 GGCCUCAGUUGGCCUUCGAGUUUGCCUGAAU--AUCAAUUCAUUAAAGUUAAUGAAUGUAAUGAGCAGAGCGCAC----ACACACUCAAUCAUAAGCGG---UGUGCCUGGUUUAUUCAU ((((......))))...((((..(((((..(--((..((((((((....))))))))...)))..)).((.((((----((.(.((........)).))---))))))))))..)))).. ( -32.10) >DroSim_CAF1 13210 113 - 1 GGCCUCAUUUGGCCUUCGAGUUUGCCUGAAU--AUCAAUUCAUUAAAGUUAAUGAAUGUAAUGAGCAGAGCACACA--CACACACUCAAUCAAAAGCGG---UGUGCCUGGCUUAUUCAU ((((......))))...((((..(((.....--....((((((((....)))))))).........((.((((((.--(.(..............).))---))))))))))..)))).. ( -30.84) >DroYak_CAF1 12983 105 - 1 GGCCUCGGAUGGCCUUCGAGUUUGCCUGAAU--AUCAAUUCAUUAAACUUAAUGAAUGUAAUGAGCAGAGCAC--------ACACUCAAUCACAAGCGG-----UGCCUGGUUUAUUCAU ((((......))))...(((((((..(((((--....))))).))))))).(((((((....((.(((.((((--------.(.((........)).))-----)))))).))))))))) ( -28.50) >consensus GGCCUCAGAUGGCCUUCGAGUUUGCCUGAAU__AUCAAUUCAUUAAAGUUAAUGAAUGUAAUGAGCAGAGCACAC____ACACACUCAAUCAAAAGCGG___UGUGCCUGGUUUAUUCAU ((((......))))...((((..(((...........((((((((....)))))))).........((.(((((............................))))))))))..)))).. (-24.15 = -24.09 + -0.06)


| Location | 8,534,280 – 8,534,395 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.82 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -22.40 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8534280 115 + 20766785 UGUGUGUGUGCUCUGCUCAUUACAUUCAUUAACUUUAAUGAAUUGAUAUAUUCAGGCAAACUCGAAGGCCAUCAAAGGCCUACGACUUAUCUGAAUAUUGGCUGUGUGUAUAAUA----- ....((((..(.(.(((.((((.((((((((....))))))))))))((((((((..((..(((.(((((......))))).))).))..)))))))).))).).)..))))...----- ( -34.00) >DroSec_CAF1 9783 109 + 1 U----GUGCGCUCUGCUCAUUACAUUCAUUAACUUUAAUGAAUUGAU--AUUCAGGCAAACUCGAAGGCCAACUGAGGCCUACGACUUAUCUGAAUAUUGGCUGUGUGCUUAAUA----- .----(..(((...(((......((((((((....)))))))).(((--((((((..((..(((.(((((......))))).))).))..)))))))))))).)))..)......----- ( -36.20) >DroSim_CAF1 13247 111 + 1 UG--UGUGUGCUCUGCUCAUUACAUUCAUUAACUUUAAUGAAUUGAU--AUUCAGGCAAACUCGAAGGCCAAAUGAGGCCUACGACUUAUCUGAAUAUUGGCUGUGUGUAUAGUA----- ..--((((..(.(.(((......((((((((....)))))))).(((--((((((..((..(((.(((((......))))).))).))..)))))))))))).).)..))))...----- ( -34.80) >DroYak_CAF1 13017 109 + 1 -------GUGCUCUGCUCAUUACAUUCAUUAAGUUUAAUGAAUUGAU--AUUCAGGCAAACUCGAAGGCCAUCCGAGGCCUACGACUUAUAAAAAAAGUG--UAUGUGUAUAAUAAUUAC -------.(((.(((...((((.((((((((....))))))))))))--...))))))...(((.(((((......))))).)))...............--.................. ( -24.00) >consensus U____GUGUGCUCUGCUCAUUACAUUCAUUAACUUUAAUGAAUUGAU__AUUCAGGCAAACUCGAAGGCCAACUGAGGCCUACGACUUAUCUGAAUAUUGGCUGUGUGUAUAAUA_____ .........((.(((...((((.((((((((....)))))))))))).....)))))....(((.(((((......))))).)))................................... (-22.40 = -22.40 + 0.00)

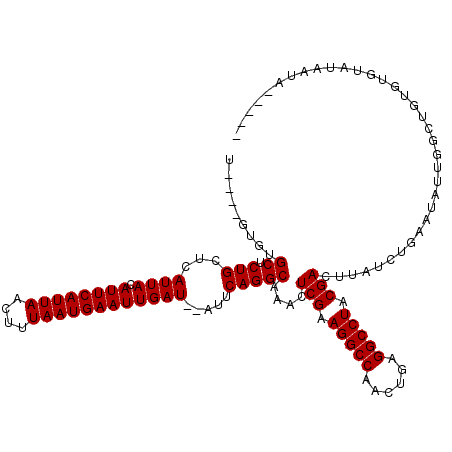
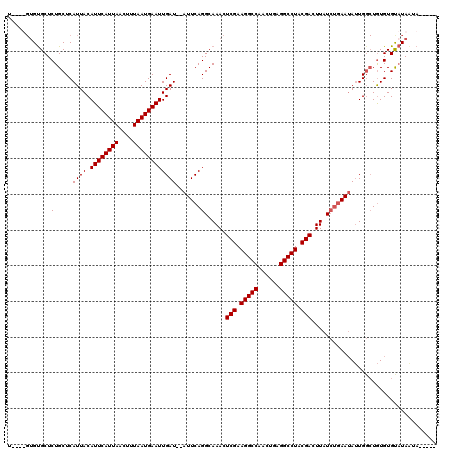
| Location | 8,534,280 – 8,534,395 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.82 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -25.34 |
| Energy contribution | -25.16 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8534280 115 - 20766785 -----UAUUAUACACACAGCCAAUAUUCAGAUAAGUCGUAGGCCUUUGAUGGCCUUCGAGUUUGCCUGAAUAUAUCAAUUCAUUAAAGUUAAUGAAUGUAAUGAGCAGAGCACACACACA -----.............((((((((((((.(((((((.(((((......))))).))).)))).))))))))....((((((((....))))))))....)).)).............. ( -30.80) >DroSec_CAF1 9783 109 - 1 -----UAUUAAGCACACAGCCAAUAUUCAGAUAAGUCGUAGGCCUCAGUUGGCCUUCGAGUUUGCCUGAAU--AUCAAUUCAUUAAAGUUAAUGAAUGUAAUGAGCAGAGCGCAC----A -----......((.(.(.((((((((((((.(((((((.(((((......))))).))).)))).))))))--))..((((((((....))))))))....)).)).).).))..----. ( -32.50) >DroSim_CAF1 13247 111 - 1 -----UACUAUACACACAGCCAAUAUUCAGAUAAGUCGUAGGCCUCAUUUGGCCUUCGAGUUUGCCUGAAU--AUCAAUUCAUUAAAGUUAAUGAAUGUAAUGAGCAGAGCACACA--CA -----.............((((((((((((.(((((((.(((((......))))).))).)))).))))))--))..((((((((....))))))))....)).))..........--.. ( -30.20) >DroYak_CAF1 13017 109 - 1 GUAAUUAUUAUACACAUA--CACUUUUUUUAUAAGUCGUAGGCCUCGGAUGGCCUUCGAGUUUGCCUGAAU--AUCAAUUCAUUAAACUUAAUGAAUGUAAUGAGCAGAGCAC------- ..................--............((.(((.(((((......))))).))).))((((((..(--((..((((((((....))))))))...)))..))).))).------- ( -26.20) >consensus _____UAUUAUACACACAGCCAAUAUUCAGAUAAGUCGUAGGCCUCAGAUGGCCUUCGAGUUUGCCUGAAU__AUCAAUUCAUUAAAGUUAAUGAAUGUAAUGAGCAGAGCACAC____A ................................((.(((.(((((......))))).))).))((((((.........((((((((....))))))))........))).)))........ (-25.34 = -25.16 + -0.19)

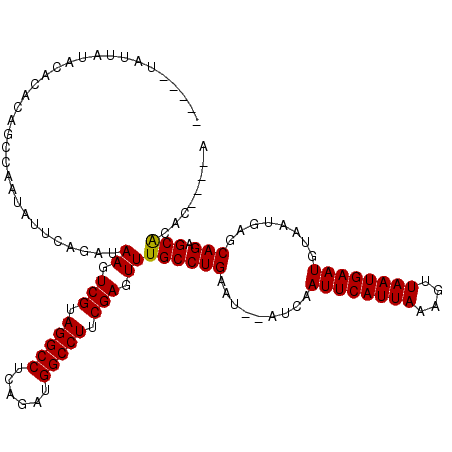
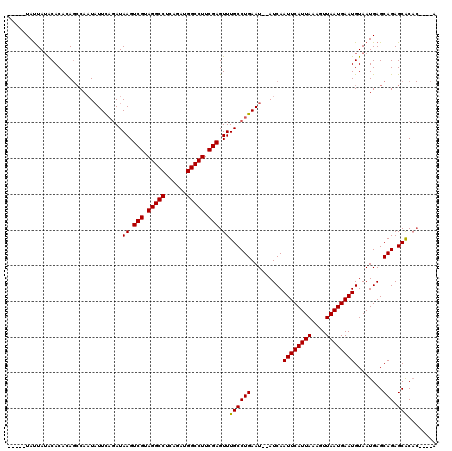
| Location | 8,534,320 – 8,534,432 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.78 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -21.23 |
| Energy contribution | -22.23 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
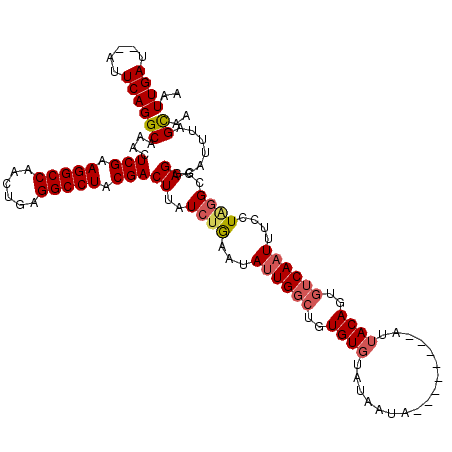
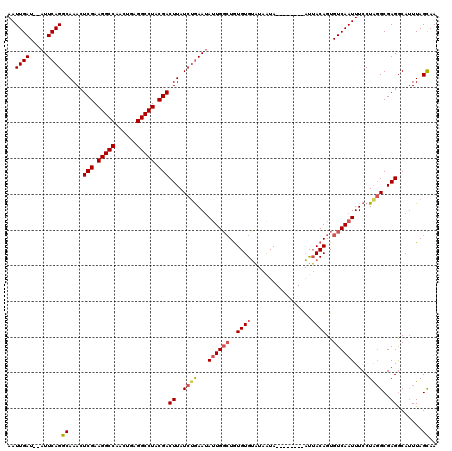
>2R_DroMel_CAF1 8534320 112 + 20766785 AAUUGAUAUAUUCAGGCAAACUCGAAGGCCAUCAAAGGCCUACGACUUAUCUGAAUAUUGGCUGUGUGUAUAAUA--------AUUACAGUGUCAAUUUCCUGGGCGAGGCAUUUAGUAA (((((((((((((((..((..(((.(((((......))))).))).))..))))))))..((((((.........--------..))))))))))))).(((.....))).......... ( -30.70) >DroSec_CAF1 9819 110 + 1 AAUUGAU--AUUCAGGCAAACUCGAAGGCCAACUGAGGCCUACGACUUAUCUGAAUAUUGGCUGUGUGCUUAAUA--------GCAACAGUGUCAAUUUCCUAGGCGAGACAUUUAGCAA (((((((--((((((..((..(((.(((((......))))).))).))..))))))....(((((.((((....)--------)))))))))))))))......((..........)).. ( -33.10) >DroSim_CAF1 13285 110 + 1 AAUUGAU--AUUCAGGCAAACUCGAAGGCCAAAUGAGGCCUACGACUUAUCUGAAUAUUGGCUGUGUGUAUAGUA--------AUUACAGUGUCAAUUUCCUAGGCGAGGCAUUUAGCAA ....(((--((((((..((..(((.(((((......))))).))).))..))))))))).((((.((((...((.--------.....(((....)))......))...)))).)))).. ( -31.70) >DroYak_CAF1 13050 116 + 1 AAUUGAU--AUUCAGGCAAACUCGAAGGCCAUCCGAGGCCUACGACUUAUAAAAAAAGUG--UAUGUGUAUAAUAAUUACAUAAUUACAGUGUCAAUUUCCUACGCGAGGCUAUUAGCAA (((((((--(((.........(((.(((((......))))).)))............(((--..((((((.......))))))..))))))))))))).....(....)((.....)).. ( -26.40) >consensus AAUUGAU__AUUCAGGCAAACUCGAAGGCCAACUGAGGCCUACGACUUAUCUGAAUAUUGGCUGUGUGUAUAAUA________AUUACAGUGUCAAUUUCCUAGGCGAGGCAUUUAGCAA ..((((.....))))((....(((.(((((......))))).)))((..((((...((((((..((((.................))))..))))))....))))..)).......)).. (-21.23 = -22.23 + 1.00)

| Location | 8,534,320 – 8,534,432 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.78 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -20.87 |
| Energy contribution | -20.69 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
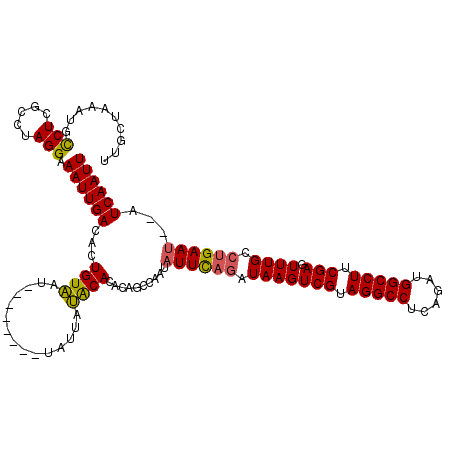
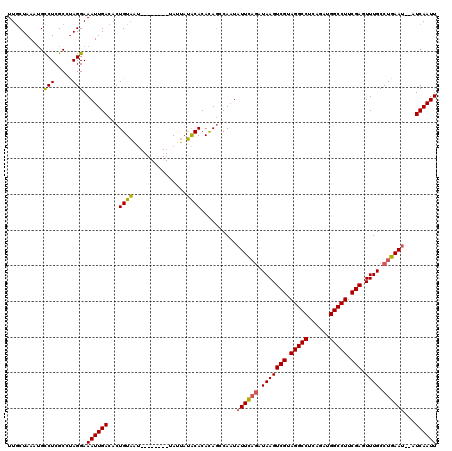
>2R_DroMel_CAF1 8534320 112 - 20766785 UUACUAAAUGCCUCGCCCAGGAAAUUGACACUGUAAU--------UAUUAUACACACAGCCAAUAUUCAGAUAAGUCGUAGGCCUUUGAUGGCCUUCGAGUUUGCCUGAAUAUAUCAAUU ..........(((.....))).((((((..((((...--------..........))))...((((((((.(((((((.(((((......))))).))).)))).)))))))).)))))) ( -27.82) >DroSec_CAF1 9819 110 - 1 UUGCUAAAUGUCUCGCCUAGGAAAUUGACACUGUUGC--------UAUUAAGCACACAGCCAAUAUUCAGAUAAGUCGUAGGCCUCAGUUGGCCUUCGAGUUUGCCUGAAU--AUCAAUU ........((((...(....).....))))(((((((--------(....)))).))))...((((((((.(((((((.(((((......))))).))).)))).))))))--))..... ( -32.00) >DroSim_CAF1 13285 110 - 1 UUGCUAAAUGCCUCGCCUAGGAAAUUGACACUGUAAU--------UACUAUACACACAGCCAAUAUUCAGAUAAGUCGUAGGCCUCAUUUGGCCUUCGAGUUUGCCUGAAU--AUCAAUU ..........(((.....))).((((((..((((...--------..........))))....(((((((.(((((((.(((((......))))).))).)))).))))))--))))))) ( -26.52) >DroYak_CAF1 13050 116 - 1 UUGCUAAUAGCCUCGCGUAGGAAAUUGACACUGUAAUUAUGUAAUUAUUAUACACAUA--CACUUUUUUUAUAAGUCGUAGGCCUCGGAUGGCCUUCGAGUUUGCCUGAAU--AUCAAUU ..((.....))......((((.....(((..((((....((((.......))))..))--)).............(((.(((((......))))).))))))..))))...--....... ( -19.60) >consensus UUGCUAAAUGCCUCGCCUAGGAAAUUGACACUGUAAU________UAUUAUACACACAGCCAAUAUUCAGAUAAGUCGUAGGCCUCAGAUGGCCUUCGAGUUUGCCUGAAU__AUCAAUU ..........(((.....))).((((((...((((...............))))..........((((((.(((((((.(((((......))))).))).)))).))))))...)))))) (-20.87 = -20.69 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:03 2006