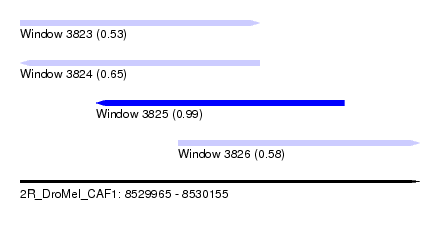
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,529,965 – 8,530,155 |
| Length | 190 |
| Max. P | 0.987909 |

| Location | 8,529,965 – 8,530,079 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.59 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -15.84 |
| Energy contribution | -15.73 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8529965 114 + 20766785 AUUAUGAGUUAGCAUAAUCGCUUGAAGAGAAUUUUUAAGUGAAAUGGCACUGUUUUGCAUGAAUUUCAAAAUAUUUCUGCCUGAUUUCGGCAGACAUUUUAUUGCGAAAUU-CAU .(((((......)))))(((((((((((...)))))))))))....(((......)))(((((((((..((((..((((((.......)))))).....))))..))))))-))) ( -30.50) >DroSec_CAF1 5490 101 + 1 AU--------------AUCGUUUAAAGAGAAUAUUUAAGUGAAAUUUCACUGUUUUGCAUUAAUUUCGAAAUAUUUCAGCCUGAUUUCGGCAGACAUUUUAUUGCAAAAUUCCAU ..--------------..........(.((((.....(((((....)))))..((((((.(((..(((((....))).(((.......))).))....))).))))))))))).. ( -16.70) >DroSim_CAF1 8740 101 + 1 AU--------------AUCGUUUGAAGAGAAUAUUUAAGUGAAAUUUCACUGUUUCGCAUCAAUUUCAAAAUAUUUCUGCCUGAUUUCGGCAGACAUUUUAUUGCAAAAUUCCAU ..--------------....((((...(((((......(((((((......))))))).................((((((.......)))))).)))))....))))....... ( -18.80) >consensus AU______________AUCGUUUGAAGAGAAUAUUUAAGUGAAAUUUCACUGUUUUGCAUCAAUUUCAAAAUAUUUCUGCCUGAUUUCGGCAGACAUUUUAUUGCAAAAUUCCAU .................(((((((((.......))))))))).........((((((((........(((((...((((((.......)))))).)))))..))))))))..... (-15.84 = -15.73 + -0.10)


| Location | 8,529,965 – 8,530,079 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.59 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -15.32 |
| Energy contribution | -16.77 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8529965 114 - 20766785 AUG-AAUUUCGCAAUAAAAUGUCUGCCGAAAUCAGGCAGAAAUAUUUUGAAAUUCAUGCAAAACAGUGCCAUUUCACUUAAAAAUUCUCUUCAAGCGAUUAUGCUAACUCAUAAU (((-(((((((.((((.....((((((.......))))))..)))).)))))))))).......((((......))))...............(((......))).......... ( -23.70) >DroSec_CAF1 5490 101 - 1 AUGGAAUUUUGCAAUAAAAUGUCUGCCGAAAUCAGGCUGAAAUAUUUCGAAAUUAAUGCAAAACAGUGAAAUUUCACUUAAAUAUUCUCUUUAAACGAU--------------AU ..(((((((((((.(((.......(((.......)))((((....))))...))).))))))).(((((....)))))......))))...........--------------.. ( -15.50) >DroSim_CAF1 8740 101 - 1 AUGGAAUUUUGCAAUAAAAUGUCUGCCGAAAUCAGGCAGAAAUAUUUUGAAAUUGAUGCGAAACAGUGAAAUUUCACUUAAAUAUUCUCUUCAAACGAU--------------AU .((((((((..(..(((((((((((((.......))))))..))))))).......((.....)))..)))))))).......................--------------.. ( -20.60) >consensus AUGGAAUUUUGCAAUAAAAUGUCUGCCGAAAUCAGGCAGAAAUAUUUUGAAAUUAAUGCAAAACAGUGAAAUUUCACUUAAAUAUUCUCUUCAAACGAU______________AU .(((((((((((..(((((((((((((.......))))))..))))))).......((.....)))))))))))))....................................... (-15.32 = -16.77 + 1.45)

| Location | 8,530,001 – 8,530,119 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -14.88 |
| Energy contribution | -17.00 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8530001 118 - 20766785 ACACUGAAUCAUUAACCACAUUUUGAAAAAUAUUCAUGGAAUG-AAUUUCGCAAUAAAAUGUCUGCCGAAAUCAGGCAGAAAUAUUUUGAAAUUCAUGCAAAACAGUGCCAUUUCACUU .(((((.........(((.....((((.....))))))).(((-(((((((.((((.....((((((.......))))))..)))).))))))))))......)))))........... ( -29.40) >DroSec_CAF1 5512 119 - 1 ACACUGAAUCAUUAACCACAUUGUGAACAAUAUUCAUGGAAUGGAAUUUUGCAAUAAAAUGUCUGCCGAAAUCAGGCUGAAAUAUUUCGAAAUUAAUGCAAAACAGUGAAAUUUCACUU .(((((.........(((..(..((((.....))))..)..)))..(((((((.(((.......(((.......)))((((....))))...))).))))))))))))........... ( -25.10) >DroSim_CAF1 8762 119 - 1 ACACUGAAUCAUUAACCACAUUGUGAACAACAUUCAUGGAAUGGAAUUUUGCAAUAAAAUGUCUGCCGAAAUCAGGCAGAAAUAUUUUGAAAUUGAUGCGAAACAGUGAAAUUUCACUU .(((((.........(((..(..((((.....))))..)..)))..(((((((.(((((((((((((.......))))))..))))))).......))))))))))))........... ( -29.20) >DroYak_CAF1 8514 92 - 1 ACACUGAAUCAAAAACCACAUUGUGAAUAAUAUUUAUGGAAUGGAAUUUCGCAUUAAAAUGUCCGCCAAAAUCAGGCAAAGAUAUUUUGUAA--------------------------- .....................((((((...(((((...)))))....)))))).(((((((((.(((.......)))...)))))))))...--------------------------- ( -14.90) >consensus ACACUGAAUCAUUAACCACAUUGUGAACAAUAUUCAUGGAAUGGAAUUUCGCAAUAAAAUGUCUGCCGAAAUCAGGCAGAAAUAUUUUGAAAUU_AUGCAAAACAGUGAAAUUUCACUU .(((((.............((((((((...(((((...)))))....))))))))((((((((((((.......))))))..))))))...............)))))........... (-14.88 = -17.00 + 2.12)

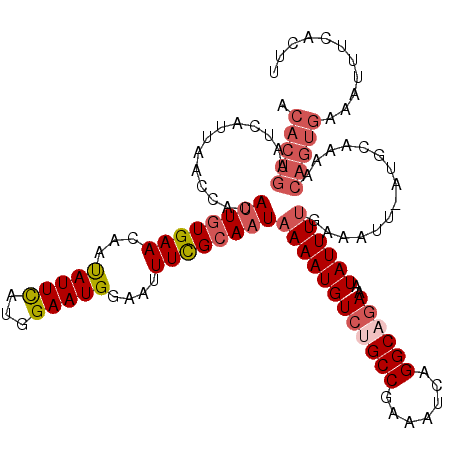
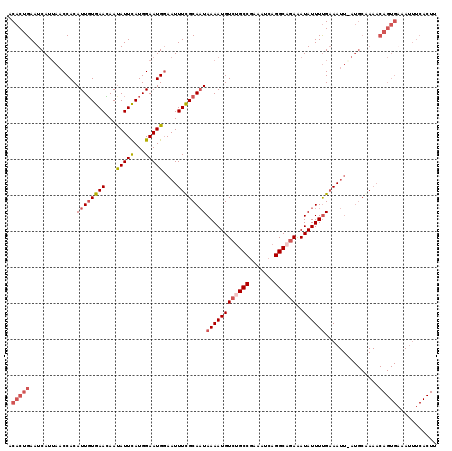
| Location | 8,530,040 – 8,530,155 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.95 |
| Mean single sequence MFE | -20.91 |
| Consensus MFE | -19.08 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8530040 115 + 20766785 UCUGCCUGAUUUCGGCAGACAUUUUAUUGCGAAAUU-CAUUCCAUGAAUAUUUUUCAAAAUGUGGUUAAUGAUUCAGUGUCGCGCCUCCAUUUUGCAUAUUCCCAUCCCUUU-UUUU--- ((((((.......)))))).................-........((((((....(((((((.(((...((((.....)))).)))..))))))).))))))..........-....--- ( -20.80) >DroSec_CAF1 5551 116 + 1 UCAGCCUGAUUUCGGCAGACAUUUUAUUGCAAAAUUCCAUUCCAUGAAUAUUGUUCACAAUGUGGUUAAUGAUUCAGUGUCGCGCCUCCAUUUUGCAUAUUCCCAUCCCUUU-UUUU--- ...((..(((...(((.((((((..(((........((((....(((((...)))))....)))).....)))..))))))..)))...)))..))................-....--- ( -19.22) >DroSim_CAF1 8801 117 + 1 UCUGCCUGAUUUCGGCAGACAUUUUAUUGCAAAAUUCCAUUCCAUGAAUGUUGUUCACAAUGUGGUUAAUGAUUCAGUGUCGCGCCUCCAUUUUGCAUAUUCCCAUCCCUUUUUUUU--- ((((((.......))))))........((((((((.((((....(((((...)))))....)))).....(((.....)))........))))))))....................--- ( -21.60) >DroYak_CAF1 8526 119 + 1 UUUGCCUGAUUUUGGCGGACAUUUUAAUGCGAAAUUCCAUUCCAUAAAUAUUAUUCACAAUGUGGUUUUUGAUUCAGUGUCGCGCCUCCAUUUUGCAUUUUCCCAUCCCUUU-UUUUUUG ..(((..(((...((((((((((..(((.(((((..((((.....................)))).)))))))).)))))).))))...)))..)))...............-....... ( -22.00) >consensus UCUGCCUGAUUUCGGCAGACAUUUUAUUGCAAAAUUCCAUUCCAUGAAUAUUGUUCACAAUGUGGUUAAUGAUUCAGUGUCGCGCCUCCAUUUUGCAUAUUCCCAUCCCUUU_UUUU___ ((((((.......))))))........((((((((............((((((....))))))(((...((((.....)))).)))...))))))))....................... (-19.08 = -18.57 + -0.50)

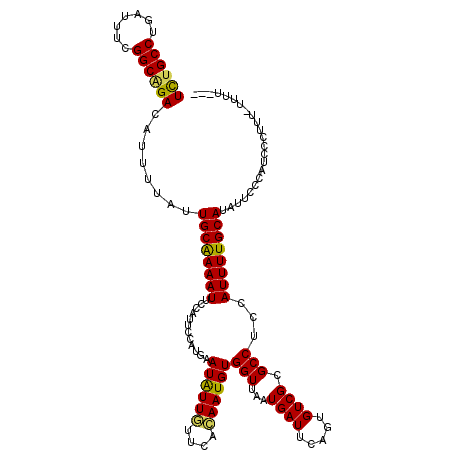

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:50 2006