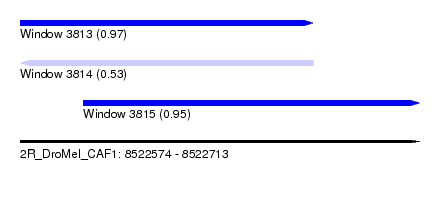
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,522,574 – 8,522,713 |
| Length | 139 |
| Max. P | 0.969573 |

| Location | 8,522,574 – 8,522,676 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -30.03 |
| Energy contribution | -30.03 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8522574 102 + 20766785 GCC-CGAUACCCGAU---ACCCGAU--------------ACACACUCACGUUCCUCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUUGAGCGAGCGUGUCAGGCAUAAAGAUA (((-.(((((.((..---...))..--------------..........((((((((..(.((((.(((((((.....))))))))))).)..)))).))))))))).)))......... ( -33.90) >DroSim_CAF1 908 98 + 1 GCCCCGAUACCCGAU---AC-------------------ACACACUCACGUUCCUCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUUGAGCGAGCGUGUCAGGCAUAAAGAUA (((.((.....))..---..-------------------.......(((((((((((..(.((((.(((((((.....))))))))))).)..)))).)))))))...)))......... ( -32.60) >DroYak_CAF1 906 119 + 1 GCC-AGAUACCCGACCCGACCCGAUAUACUCGUACACACACUCACUCACGUUCCUCAGCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUUGAGCGAGCGUGUCAGGCAUAAAGAUA (((-.................(((.....)))..............((((((((((((.(.((((.(((((((.....))))))))))).).))))).)))))))...)))......... ( -34.70) >consensus GCC_CGAUACCCGAU___ACCCGAU______________ACACACUCACGUUCCUCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUUGAGCGAGCGUGUCAGGCAUAAAGAUA (((..(.....)..................................(((((((((((..(.((((.(((((((.....))))))))))).)..)))).)))))))...)))......... (-30.03 = -30.03 + 0.00)

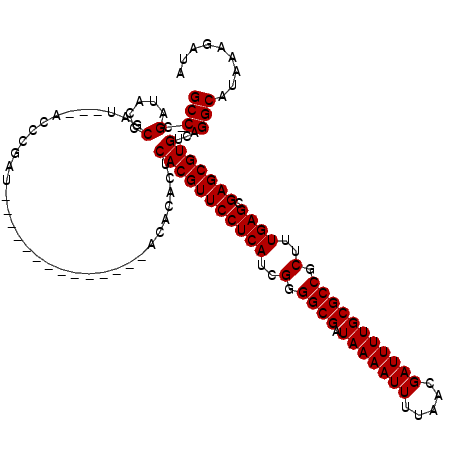
| Location | 8,522,574 – 8,522,676 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -25.49 |
| Energy contribution | -25.83 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8522574 102 - 20766785 UAUCUUUAUGCCUGACACGCUCGCUCAAAGCGGCGCAAAAUCGUUAAAAUUUUAUCGCCCCGAUGAGGAACGUGAGUGUGU--------------AUCGGGU---AUCGGGUAUCG-GGC ...((..((((((((..(((.(((.....)))))).......................(((((((....((....))...)--------------)))))).---.))))))))..-)). ( -33.80) >DroSim_CAF1 908 98 - 1 UAUCUUUAUGCCUGACACGCUCGCUCAAAGCGGCGCAAAAUCGUUAAAAUUUUAUCGCCCCGAUGAGGAACGUGAGUGUGU-------------------GU---AUCGGGUAUCGGGGC ..(((..((((((((.((((.((((((....((((.(((((.......)))))..))))((.....))....)))))).))-------------------))---.))))))))..))). ( -34.70) >DroYak_CAF1 906 119 - 1 UAUCUUUAUGCCUGACACGCUCGCUCAAAGCGGCGCAAAAUCGUUAAAAUUUUAUCGCCCCGCUGAGGAACGUGAGUGAGUGUGUGUACGAGUAUAUCGGGUCGGGUCGGGUAUCU-GGC ...((..((((((((((((((((((((.(((((.(((((((.......)))))...)).)))))........))))))))))).....(((.(......).))).)))))))))..-)). ( -41.80) >consensus UAUCUUUAUGCCUGACACGCUCGCUCAAAGCGGCGCAAAAUCGUUAAAAUUUUAUCGCCCCGAUGAGGAACGUGAGUGUGU______________AUCGGGU___AUCGGGUAUCG_GGC .......(((((((((((((((((.......((((.(((((.......)))))..))))((.....))...)))))))))..........................))))))))...... (-25.49 = -25.83 + 0.33)

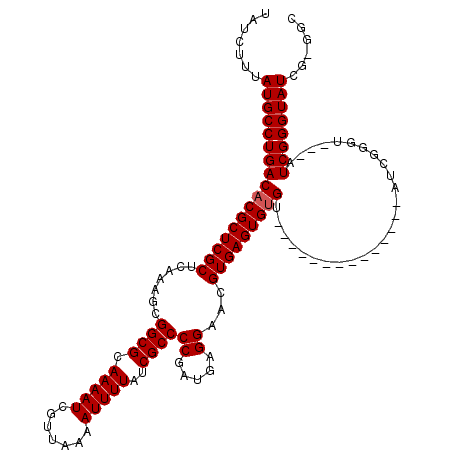
| Location | 8,522,596 – 8,522,713 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.20 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8522596 117 + 20766785 CACACUCACGUUCCUCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUUGAGCGAGCGUGUCAGGCAUAAAGAUACAAAACAAUACUUGGGCAUAUA---UAAACAAAAGCAAAC ......(((((((((((..(.((((.(((((((.....))))))))))).)..)))).)))))))....((...(((((........)).)))..)).....---............... ( -30.00) >DroSim_CAF1 926 117 + 1 CACACUCACGUUCCUCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUUGAGCGAGCGUGUCAGGCAUAAAGAUACAAAACAAUACUUGGGCAUAUA---UAAACAAAAGCAAAC ......(((((((((((..(.((((.(((((((.....))))))))))).)..)))).)))))))....((...(((((........)).)))..)).....---............... ( -30.00) >DroYak_CAF1 945 120 + 1 CUCACUCACGUUCCUCAGCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUUGAGCGAGCGUGUCAGGCAUAAAGAUACAAAACAAUACUUGGGCAUAUACAAUAAACAAAAGCAAAC ......((((((((((((.(.((((.(((((((.....))))))))))).).))))).)))))))....((...(((((........)).)))..))....................... ( -30.50) >consensus CACACUCACGUUCCUCAUCGGGGCGAUAAAAUUUUAACGAUUUUGCGCCGCUUUGAGCGAGCGUGUCAGGCAUAAAGAUACAAAACAAUACUUGGGCAUAUA___UAAACAAAAGCAAAC ......(((((((((((..(.((((.(((((((.....))))))))))).)..)))).)))))))....((...(((((........)).)))..))....................... (-29.80 = -29.80 + 0.00)

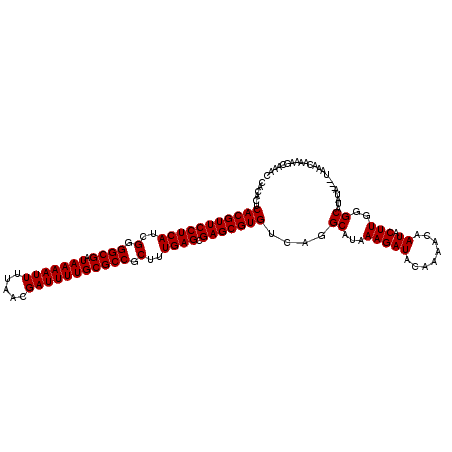
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:40 2006