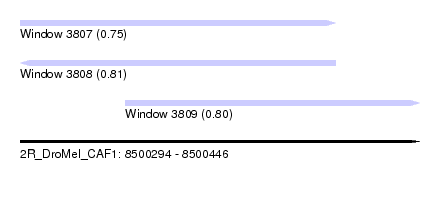
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,500,294 – 8,500,446 |
| Length | 152 |
| Max. P | 0.806356 |

| Location | 8,500,294 – 8,500,414 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.97 |
| Mean single sequence MFE | -39.35 |
| Consensus MFE | -31.31 |
| Energy contribution | -32.00 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8500294 120 + 20766785 GACACUCUUUCGCUUUCGAGGAGAUCUAUCUCCUUAGAUCCAUUAGUUUGGACCUAUCGAGGCAUCACUGUAGUCUUGAGCCGCACAGUGAAAAGACGAUGGAGCCAAGCUAAAGUCGUG (((........(..((..((((((....))))))..))..).(((((((((..((((((.....(((((((.((........))))))))).....))))))..))))))))).)))... ( -42.30) >DroSec_CAF1 12859 120 + 1 GACACUCUUUCGCUUUGGAGGAGAUCUAUCUCCUUAGAUCCCAAGAUUUGAACCUAUCGAGGCUUCACUGUAGUCUUGAGCCGCUCAGUGAAAAGACGAUGGAGCCAAGCCAAAGUCGUG ..(((.((((.((((.((.((.((((((......))))))))...........((((((....(((((((.(((........))))))))))....))))))..)))))).))))..))) ( -37.20) >DroSim_CAF1 12744 120 + 1 GACACUCUUUCGCUUUGGAGGAGAUCUAUCUCCUUAGAUCCCUAGGUUUGGACCUAGCGAGGCAUCACUGUAGUCUUGAGCCGCUCAGUGAAAAGACGAUGGAGCCAAGCCAAAGUCGUG ..(((......(((((((((((((....)))))))......((((((....)))))))))))).((((((.(((........)))))))))...(((..(((.......)))..)))))) ( -40.00) >DroEre_CAF1 10452 120 + 1 GGCACUCUUUCGCCUUAGAAGGGACUUAGCUCCUAAGAUCCAUUAGCUUGGACCUAUCGAGGCAUCACUGUAGUCUUGAGCCGCUCAGUGAAAAGACGAUGGAGCCAAGCCAAAGUCGUG (((.((((.(((((((.((.(((.(((((...))))).((((......))))))).))))))).((((((.(((........)))))))))......)).))))....)))......... ( -37.90) >consensus GACACUCUUUCGCUUUGGAGGAGAUCUAUCUCCUUAGAUCCAUAAGUUUGGACCUAUCGAGGCAUCACUGUAGUCUUGAGCCGCUCAGUGAAAAGACGAUGGAGCCAAGCCAAAGUCGUG (((........(..((..(((((......)))))..))..)...(((((((..((((((.....((((((.(((........))))))))).....))))))..)))))))...)))... (-31.31 = -32.00 + 0.69)

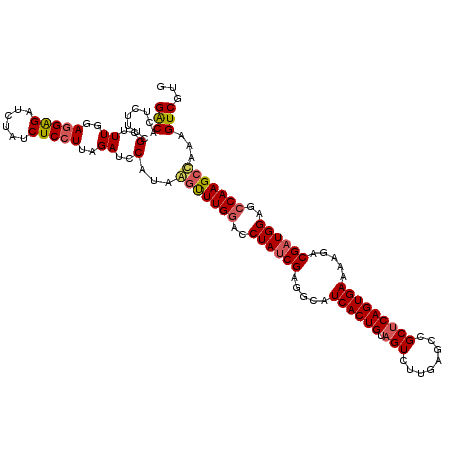
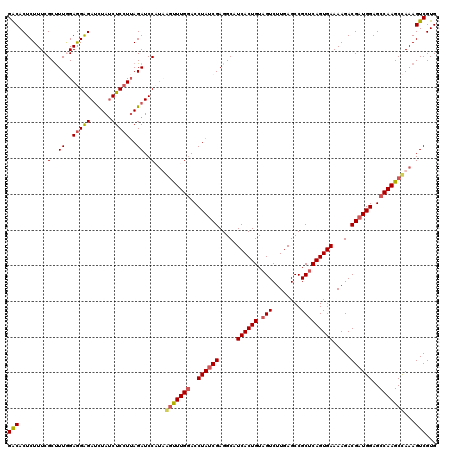
| Location | 8,500,294 – 8,500,414 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.97 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -30.69 |
| Energy contribution | -30.82 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8500294 120 - 20766785 CACGACUUUAGCUUGGCUCCAUCGUCUUUUCACUGUGCGGCUCAAGACUACAGUGAUGCCUCGAUAGGUCCAAACUAAUGGAUCUAAGGAGAUAGAUCUCCUCGAAAGCGAAAGAGUGUC (((..((((.(((((((............((((((((.(........))))))))).)))((((((((((((......)))))))).(((((....))))))))))))).)))).))).. ( -39.92) >DroSec_CAF1 12859 120 - 1 CACGACUUUGGCUUGGCUCCAUCGUCUUUUCACUGAGCGGCUCAAGACUACAGUGAAGCCUCGAUAGGUUCAAAUCUUGGGAUCUAAGGAGAUAGAUCUCCUCCAAAGCGAAAGAGUGUC (((..((((.((((((.....(((...(((((((((((.......).)).))))))))...)))(((((((........)))))))((((((....)))))))).)))).)))).))).. ( -36.70) >DroSim_CAF1 12744 120 - 1 CACGACUUUGGCUUGGCUCCAUCGUCUUUUCACUGAGCGGCUCAAGACUACAGUGAUGCCUCGCUAGGUCCAAACCUAGGGAUCUAAGGAGAUAGAUCUCCUCCAAAGCGAAAGAGUGUC (((..((((.((((((.(((..((.....(((((((((.......).)).)))))).....))((((((....)))))))))....((((((....)))))))).)))).)))).))).. ( -38.10) >DroEre_CAF1 10452 120 - 1 CACGACUUUGGCUUGGCUCCAUCGUCUUUUCACUGAGCGGCUCAAGACUACAGUGAUGCCUCGAUAGGUCCAAGCUAAUGGAUCUUAGGAGCUAAGUCCCUUCUAAGGCGAAAGAGUGCC (((..(((((((((((((((((((.....(((((((((.......).)).)))))).....))))(((((((......)))))))..)))))))))))(((....)))..)))).))).. ( -43.20) >consensus CACGACUUUGGCUUGGCUCCAUCGUCUUUUCACUGAGCGGCUCAAGACUACAGUGAUGCCUCGAUAGGUCCAAACCAAGGGAUCUAAGGAGAUAGAUCUCCUCCAAAGCGAAAGAGUGUC (((..((((.((((..((((((((.....(((((((((.......).)).)))))).....))))((((((........))))))..))))..............)))).)))).))).. (-30.69 = -30.82 + 0.12)

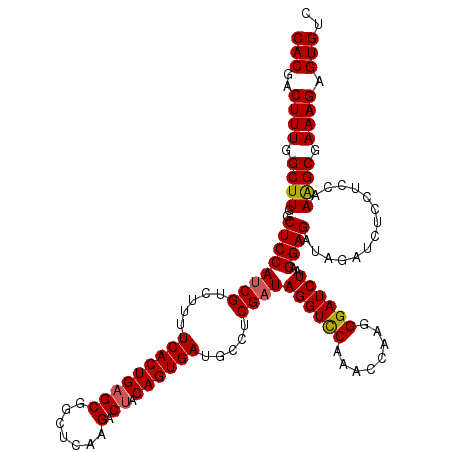
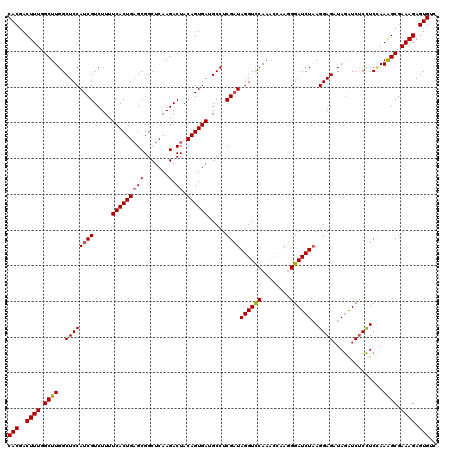
| Location | 8,500,334 – 8,500,446 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -18.31 |
| Energy contribution | -19.82 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
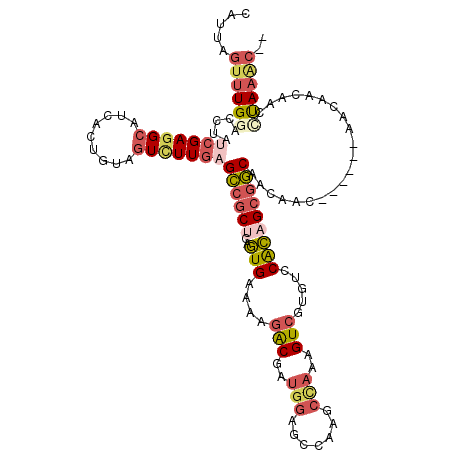
>2R_DroMel_CAF1 8500334 112 + 20766785 CAUUAGUUUGGACCUAUCGAGGCAUCACUGUAGUCUUGAGCCGCACAGUGAAAAGACGAUGGAGCCAAGCUAAAGUCGUGUCCACAGCGGCAACAAU------AACAACAACCUAAAC-- ..(((((((((..((((((.....(((((((.((........))))))))).....))))))..))))))))).(((((.......)))))......------...............-- ( -34.40) >DroSec_CAF1 12899 112 + 1 CCAAGAUUUGAACCUAUCGAGGCUUCACUGUAGUCUUGAGCCGCUCAGUGAAAAGACGAUGGAGCCAAGCCAAAGUCGUGUCCACAGCGGCAACAAC------AACAACAACCUAAAC-- .((((((((((((((....))).))))....))))))).((((((..(((....(((..(((.......)))..))).....)))))))))......------...............-- ( -30.60) >DroSim_CAF1 12784 112 + 1 CCUAGGUUUGGACCUAGCGAGGCAUCACUGUAGUCUUGAGCCGCUCAGUGAAAAGACGAUGGAGCCAAGCCAAAGUCGUGUCCACAGCGGCAACAAC------AACAACAAUCUAAAC-- .((((((....))))))((((((.........)))))).((((((..(((....(((..(((.......)))..))).....)))))))))......------...............-- ( -31.90) >DroEre_CAF1 10492 109 + 1 CAUUAGCUUGGACCUAUCGAGGCAUCACUGUAGUCUUGAGCCGCUCAGUGAAAAGACGAUGGAGCCAAGCCAAAGUCGUGUCCGCAGCGGCAA---C------AACAACAACUUAAAC-- .....((((((..((((((.....((((((.(((........))))))))).....))))))..))))))....(((((.......)))))..---.------...............-- ( -33.10) >DroYak_CAF1 13458 112 + 1 CAUUAGUGUGGGCCUAUCGAGGCAUCACUGUAGUUUUGAGCCGCUCAGUGAAAAGACGAUGGAGCCAAGCCAAAGUCGUGUCCGCAGCGGCAAUAAC------AACAACAACUUAAAC-- ...(((((...((((....))))..)))))..(((....((((((..(.((...(((..(((.......)))..)))...)))..))))))...)))------...............-- ( -31.70) >DroAna_CAF1 9680 106 + 1 CU---GUUUG-GUUUAUGGACGCUCCACUGUAAUUUUGUGGCGCGGAUUGAACAUGCGA----------CUUAAGUCGUGUCCAAAGAUCCACUAGCCUGUGCUCCAACAACUCAAGCAA .(---(((((-(((..(((((((.((((.........)))).))(((((....((((((----------(....))))))).....)))))..........).))))..))).)))))). ( -33.30) >consensus CAUUAGUUUGGACCUAUCGAGGCAUCACUGUAGUCUUGAGCCGCUCAGUGAAAAGACGAUGGAGCCAAGCCAAAGUCGUGUCCACAGCGGCAACAAC______AACAACAACCUAAAC__ .....((((((.....(((((((.........)))))))((((((..(((....(((..(((.......)))..))).....))))))))).....................)))))).. (-18.31 = -19.82 + 1.51)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:34 2006