
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,497,401 – 8,497,547 |
| Length | 146 |
| Max. P | 0.999659 |

| Location | 8,497,401 – 8,497,507 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -16.08 |
| Energy contribution | -19.00 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
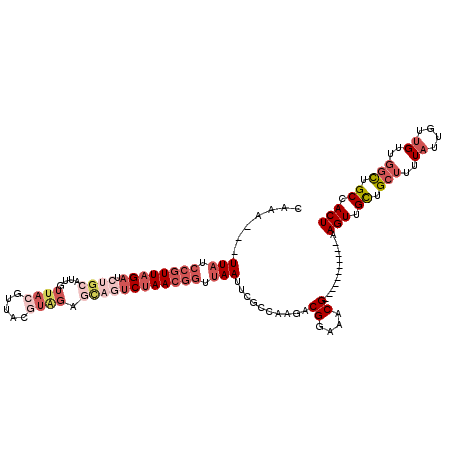
>2R_DroMel_CAF1 8497401 106 - 20766785 CAAU---UUAUCCGUUAGAUCUGCAUUGUUACGUUACGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG-----------AAGUUGCUGCUUUUAUUGUUGUUGGCUGCCACU ....---(((.((((((((.((((....(((((...))))).)))))))))))).)))...(((((((((....)(-----------((((....)))))....))).)))))....... ( -32.30) >DroSec_CAF1 9716 106 - 1 CAAA---UUAUCCGUUAGAUCUGCAUUGUUACGUUACGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG-----------AAGUUGCUGCUUUUAUUGUUGUUGGUUGCCACU ..((---(((.((((((((.((((....(((((...))))).)))))))))))).))))).(((((((((....)(-----------((((....)))))....))).)))))....... ( -31.40) >DroSim_CAF1 9629 106 - 1 CAAA---UUAUCCGUUAGAUCUGCAUUGUUACGUUACGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG-----------AAGUUGCUGCUUUUAUUGUUGUUGGCUGCCACU ..((---(((.((((((((.((((....(((((...))))).)))))))))))).))))).(((((((((....)(-----------((((....)))))....))).)))))....... ( -33.90) >DroEre_CAF1 7288 90 - 1 CAAA---UUAUCCGUUAGAU----------------CGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG-----------AAGUUGCUGCUUUUAUUGUUGUUGGCUGCCACU ..((---(((.(((((((((----------------.(.....).))))))))).))))).(((((((((....)(-----------((((....)))))....))).)))))....... ( -26.90) >DroYak_CAF1 10050 106 - 1 CAAA---UUAUCCGUUAGAUCUGCAUUGUUACGUUACGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG-----------AAGUUGCUGCUUUUAUUGUUGUUGGCUGCCACU ..((---(((.((((((((.((((....(((((...))))).)))))))))))).))))).(((((((((....)(-----------((((....)))))....))).)))))....... ( -33.90) >DroAna_CAF1 7244 113 - 1 CAAAGCGUUAUCCGUUAGAUCAGCAUUGUUA-GUUUCGUGGGUUAAGCAAAAAAUAAAA---AAAAAACGGAAACGGGAACGGUGUGGAGUUGUUG---UUCUUGGAGCUUGUUGCCACU (((.((...(((.....)))..)).)))...-.....((((..(((((...........---......((....))((((((..(......)..))---))))....)))))...)))). ( -23.30) >consensus CAAA___UUAUCCGUUAGAUCUGCAUUGUUACGUUACGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG___________AAGUUGCUGCUUUUAUUGUUGUUGGCUGCCACU .......(((.((((((((.((((....((((.....)))).)))))))))))).)))..........((....))............(((.((.(((..((....))..))).)).))) (-16.08 = -19.00 + 2.92)

| Location | 8,497,434 – 8,497,547 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 92.21 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -29.43 |
| Energy contribution | -30.87 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8497434 113 + 20766785 CGUUUCCGUCUUGGCGAAUUAACCGUUAGACUGCUCUACGUAACGUAACAAUGCAGAUCUAACGGAUAAAUUGCGAACGAAGGAACCGUACCAUGACGAUACUUUCCAUGCAA ((.((((.((((.((((.(((.((((((((((((..((((...)))).....)))).)))))))).))).)))).)).)).)))).))...((((..((.....))))))... ( -33.70) >DroSec_CAF1 9749 113 + 1 CGUUUCCGUCUUGGCGAAUUAACCGUUAGACUGCUCUACGUAACGUAACAAUGCAGAUCUAACGGAUAAUUUGCGAGCGAAGGAACCGUACCAUGACGAUACUUUCCAUGCAA ((.((((.((((.((((((((.((((((((((((..((((...)))).....)))).)))))))).)))))))).)).)).)))).))...((((..((.....))))))... ( -37.10) >DroSim_CAF1 9662 113 + 1 CGUUUCCGUCUUGGCGAAUUAACCGUUAGACUGCUCUACGUAACGUAACAAUGCAGAUCUAACGGAUAAUUUGCGAGCGAAGGAACCGUACCAUGACGAUACUUUCCAUGCAA ((.((((.((((.((((((((.((((((((((((..((((...)))).....)))).)))))))).)))))))).)).)).)))).))...((((..((.....))))))... ( -37.10) >DroEre_CAF1 7321 97 + 1 CGUUUCCGUCUUGGCGAAUUAACCGUUAGACUGCUCUACG----------------AUCUAACGGAUAAUUUGCGAGCGCAGGAACCGUACCAUGACGAUUCUUUCCAUGCAA ((.((((((.((.((((((((.((((((((.((.....))----------------.)))))))).)))))))).)).)).)))).))...((((..((.....))))))... ( -29.60) >DroYak_CAF1 10083 113 + 1 CGUUUCCGUCUUGGCGAAUUAACCGUUAGACUGCUCUACGUAACGUAACAAUGCAGAUCUAACGGAUAAUUUGCGAGCAAAGGAACCGUACCAUGACGAUACUUUCAAUGCAA ((.((((...((.((((((((.((((((((((((..((((...)))).....)))).)))))))).)))))))).))....)))).))......................... ( -34.10) >consensus CGUUUCCGUCUUGGCGAAUUAACCGUUAGACUGCUCUACGUAACGUAACAAUGCAGAUCUAACGGAUAAUUUGCGAGCGAAGGAACCGUACCAUGACGAUACUUUCCAUGCAA ((.((((.((((.((((((((.((((((((((((..................)))).)))))))).)))))))).)).)).)))).))...((((..((.....))))))... (-29.43 = -30.87 + 1.44)


| Location | 8,497,434 – 8,497,547 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 92.21 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -29.74 |
| Energy contribution | -31.78 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
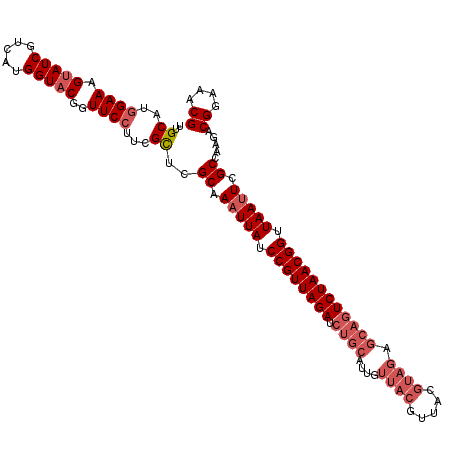
>2R_DroMel_CAF1 8497434 113 - 20766785 UUGCAUGGAAAGUAUCGUCAUGGUACGGUUCCUUCGUUCGCAAUUUAUCCGUUAGAUCUGCAUUGUUACGUUACGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG ......((((.(((((.....)))))..))))((((((.((((((...((((((((.((((....(((((...))))).))))))))))))..)))).))...)))))).... ( -35.70) >DroSec_CAF1 9749 113 - 1 UUGCAUGGAAAGUAUCGUCAUGGUACGGUUCCUUCGCUCGCAAAUUAUCCGUUAGAUCUGCAUUGUUACGUUACGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG ..((..((((.(((((.....)))))..))))...))..((.(((((.((((((((.((((....(((((...))))).)))))))))))).))))).)).....((....)) ( -38.10) >DroSim_CAF1 9662 113 - 1 UUGCAUGGAAAGUAUCGUCAUGGUACGGUUCCUUCGCUCGCAAAUUAUCCGUUAGAUCUGCAUUGUUACGUUACGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG ..((..((((.(((((.....)))))..))))...))..((.(((((.((((((((.((((....(((((...))))).)))))))))))).))))).)).....((....)) ( -38.10) >DroEre_CAF1 7321 97 - 1 UUGCAUGGAAAGAAUCGUCAUGGUACGGUUCCUGCGCUCGCAAAUUAUCCGUUAGAU----------------CGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG ..(((.(((....(((((......)))))))))))....((.(((((.(((((((((----------------.(.....).))))))))).))))).)).....((....)) ( -29.50) >DroYak_CAF1 10083 113 - 1 UUGCAUUGAAAGUAUCGUCAUGGUACGGUUCCUUUGCUCGCAAAUUAUCCGUUAGAUCUGCAUUGUUACGUUACGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG ..(((..(((.(((((.....)))))..)))...)))..((.(((((.((((((((.((((....(((((...))))).)))))))))))).))))).)).....((....)) ( -36.20) >consensus UUGCAUGGAAAGUAUCGUCAUGGUACGGUUCCUUCGCUCGCAAAUUAUCCGUUAGAUCUGCAUUGUUACGUUACGUAGAGCAGUCUAACGGUUAAUUCGCCAAGACGGAAACG ..((..((((.(((((.....)))))..))))...))..((.(((((.((((((((.((((....((((.....)))).)))))))))))).))))).)).....((....)) (-29.74 = -31.78 + 2.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:31 2006