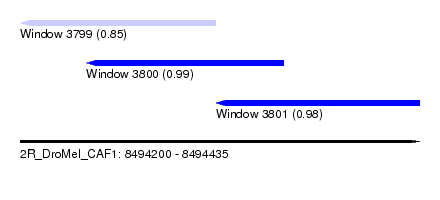
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,494,200 – 8,494,435 |
| Length | 235 |
| Max. P | 0.991310 |

| Location | 8,494,200 – 8,494,315 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.96 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8494200 115 - 20766785 UCUUCGCGCACCAACGAGGCUAUACCGAU----AUAGCUUCGGCUUAUUGCUUUCUGCGGCUCAGCGUCUGAUUUAUUUAUAAUCUUCAGCACGCUCUCUUCUCGAGUCGAUUUACAUU- ...((..(((....((((((((((....)----)))))))))((.....))....)))((((((((((((((..............)))).)))))........)))))))........- ( -31.94) >DroSec_CAF1 6551 115 - 1 UCUUCGCGCACCAACGAGGCUAUACCGAU----AUAGCUUCAGCUUAUUGCUUUCUGCGGCUCAGCGGCUGAUUUAUUUAUAAUCUUCAGCACGCUCUCUUCUCAAGUCGAUUUUCAUU- ...(((((((.....(((((((((....)----))))))))(((.....)))...))))....(((((((((..............))))).))))............)))........- ( -30.04) >DroSim_CAF1 6469 115 - 1 UCUUCGCGCACCAACGAGGCUUUACCAAU----AUAGCUUCAGCUUAUUGCUUUCUGCGGCUCAGCGGCUGAUUUAUUUAUAAUCUUCAGCACGCUCUCUUCUCAAGUCGAUUUUCAUU- ...(((((((.....((((((........----..))))))(((.....)))...))))....(((((((((..............))))).))))............)))........- ( -26.14) >DroEre_CAF1 4014 112 - 1 UUGC--CCCACCAACGAAGCUACA--GAU----AUAGCUUCAGCUUAUUGCUUUCUGCGGCUCAGCCGCUGAUUUAUUUAUAAUCUUCAGCACGCUCUCGUCUCAAGUCGAUUUUGAAUU ..((--(.((.....(((((((..--...----.)))))))(((.....)))...)).)))(((((.(((((..............)))))..))..(((.(....).)))...)))... ( -25.14) >DroYak_CAF1 6760 118 - 1 UCUC--CCCAUCAACGAAGCUACACAGAUAUAUGUAGCUUCAGCUUAUUGCUUUCUGCGGCUCAACCGCUGAUUUAUUAAUAAUCUUCAGCACGCUCUCGUCUCAAGUCGAUUUUCGUUU ....--.........(((((((((........)))))))))(((.....)))....((((.....))(((((..............)))))..))..(((.(....).)))......... ( -25.64) >consensus UCUUCGCGCACCAACGAGGCUAUACCGAU____AUAGCUUCAGCUUAUUGCUUUCUGCGGCUCAGCGGCUGAUUUAUUUAUAAUCUUCAGCACGCUCUCUUCUCAAGUCGAUUUUCAUU_ ...............((((((((..........))))))))(((.....))).....(((((.(((((((((..............))))).)))).........))))).......... (-19.56 = -20.12 + 0.56)

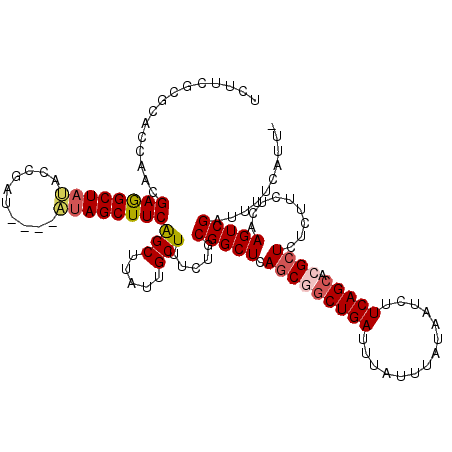
| Location | 8,494,239 – 8,494,355 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.69 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -30.74 |
| Energy contribution | -30.70 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8494239 116 - 20766785 UUCUAUCGUAAACAGCGAGGGGCGAGUGGAGCUUUAAGCUUCUUCGCGCACCAACGAGGCUAUACCGAU----AUAGCUUCGGCUUAUUGCUUUCUGCGGCUCAGCGUCUGAUUUAUUUA .......((((((((((..(((((((.(((((.....)))))))).((((....((((((((((....)----)))))))))((.....))....))))))))..)).))).)))))... ( -40.40) >DroSec_CAF1 6590 116 - 1 UUCUUUCGUAAACAGCGAGGGGCGAGUGGAGCUUUAAGCUUCUUCGCGCACCAACGAGGCUAUACCGAU----AUAGCUUCAGCUUAUUGCUUUCUGCGGCUCAGCGGCUGAUUUAUUUA .......((((((((((..(.(((((.(((((.....)))))))))).)..)...(((((((((....)----)))))))).(((..((((.....))))...))).)))).)))))... ( -39.50) >DroSim_CAF1 6508 116 - 1 UUCUUUCGUAAACAGCGAGGGGCGAGUGGAGCUUUAAGCUUCUUCGCGCACCAACGAGGCUUUACCAAU----AUAGCUUCAGCUUAUUGCUUUCUGCGGCUCAGCGGCUGAUUUAUUUA .......((((((((((..(.(((((.(((((.....)))))))))).)..)...((((((........----..)))))).(((..((((.....))))...))).)))).)))))... ( -35.60) >DroEre_CAF1 4054 112 - 1 UUCUUUCGUAAACAGCGUGGGGCGAGUGGAGCUUUAAGCUUUGC--CCCACCAACGAAGCUACA--GAU----AUAGCUUCAGCUUAUUGCUUUCUGCGGCUCAGCCGCUGAUUUAUUUA .......((((..((((((((((((....(((.....)))))))--)))))....(((((((..--...----.))))))).)))..))))..((.(((((...))))).))........ ( -41.70) >DroYak_CAF1 6800 118 - 1 UUCUUUCGUAAACAGCGAGGGGCGAUUGGAGCUUUAAGCUUCUC--CCCAUCAACGAAGCUACACAGAUAUAUGUAGCUUCAGCUUAUUGCUUUCUGCGGCUCAACCGCUGAUUUAUUAA .......((((((((((.(((.((...(((((..((((((....--.........(((((((((........)))))))))))))))..)))))...)).)))...))))).)))))... ( -38.91) >consensus UUCUUUCGUAAACAGCGAGGGGCGAGUGGAGCUUUAAGCUUCUUCGCGCACCAACGAGGCUAUACCGAU____AUAGCUUCAGCUUAUUGCUUUCUGCGGCUCAGCGGCUGAUUUAUUUA .......(((((((((..(((.((...(((((..((((((...............((((((((..........))))))))))))))..)))))...)).)))....)))).)))))... (-30.74 = -30.70 + -0.04)



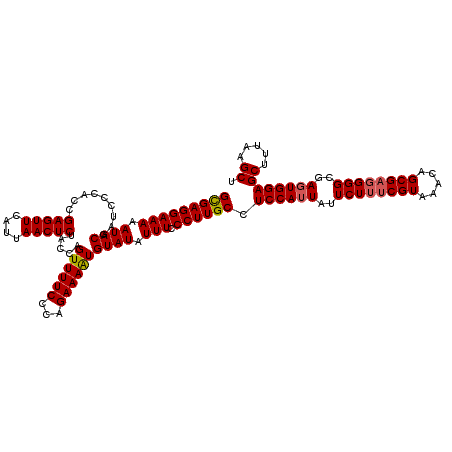
| Location | 8,494,315 – 8,494,435 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.16 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -29.88 |
| Energy contribution | -30.08 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8494315 120 - 20766785 GUGAGGAAAAAUACGAUCCCACCGAGUUCAUUAACUCUACCAGUUUUCCCAGAAAGUGUAUAUUUCCCUUGCCUCCAUUAUUCUAUCGUAAACAGCGAGGGGCGAGUGGAGCUUUAAGCU (..((((((.(((((........(((((....)))))......(((((...)))))))))).))).)))..)(((((((..(((.((((.....)))).)))..)))))))......... ( -27.90) >DroSec_CAF1 6666 120 - 1 GCGAGGAAAAAUACGAUCCCACCGAGUUCAUUAACUCGACCAGUUUUCCCAGAAAAUGUAUAUUUCCCUUGCCUCCAUUAUUCUUUCGUAAACAGCGAGGGGCGAGUGGAGCUUUAAGCU (((((((((.((((........((((((....))))))....((((((...)))))))))).))).))))))(((((((..((((((((.....))))))))..)))))))......... ( -36.90) >DroSim_CAF1 6584 120 - 1 GCGAGGAAAAAUACGAUCCCACCGAGUUCAUUAACUCGACCAGUUUUCCCAGAAAAUGUAUAUUUCCCUUGCCUCCAUUAUUCUUUCGUAAACAGCGAGGGGCGAGUGGAGCUUUAAGCU (((((((((.((((........((((((....))))))....((((((...)))))))))).))).))))))(((((((..((((((((.....))))))))..)))))))......... ( -36.90) >DroEre_CAF1 4126 119 - 1 GUGAGGAAAAAUACGAUCCCACCGAGUUCAUUAACUCUGCGAGUUUUCCCAGAAAAUGUAUAUUUCCCUUGC-UCCAUUAUUCUUUCGUAAACAGCGUGGGGCGAGUGGAGCUUUAAGCU ....(((((.((((.........(((((....))))).....((((((...)))))))))).)))))...((-((((((..(((..(((.....)))..)))..))))))))........ ( -30.70) >DroYak_CAF1 6878 119 - 1 GCGAGGAAAAAUACGAUCCCACCGAGUUCAUUAACUCUGCGAGUUUUCCCAGAAAAUGUAUAUUUCCCUUGC-UCCAUUAUUCUUUCGUAAACAGCGAGGGGCGAUUGGAGCUUUAAGCU ((..(((((.((((.........(((((....))))).....((((((...)))))))))).)))))...((-((((....((((((((.....))))))))....)))))).....)). ( -33.60) >consensus GCGAGGAAAAAUACGAUCCCACCGAGUUCAUUAACUCUACCAGUUUUCCCAGAAAAUGUAUAUUUCCCUUGCCUCCAUUAUUCUUUCGUAAACAGCGAGGGGCGAGUGGAGCUUUAAGCU (((((((((.((((.........(((((....))))).....((((((...)))))))))).))).)))))).((((((..((((((((.....))))))))..))))))((.....)). (-29.88 = -30.08 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:26 2006