
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,490,275 – 8,490,377 |
| Length | 102 |
| Max. P | 0.999095 |

| Location | 8,490,275 – 8,490,377 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -18.30 |
| Energy contribution | -19.30 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
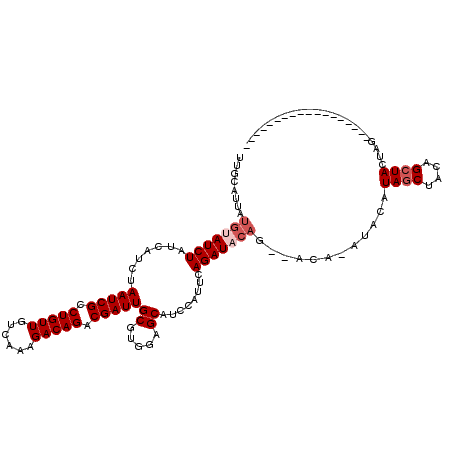
>2R_DroMel_CAF1 8490275 102 + 20766785 CUGCAUUAUGUAUCUAUCAUCUAAUCGCCUGUUGUCAAAGACAGACGAUUGCGUGGAGCAUUCAUUCAGAUACAGCUACAAAUACAUAGCUACAGCUACUAG------------------ ..((.....((((((..(((.((((((.(((((......))))).)))))).)))(((......)))))))))(((((........)))))...))......------------------ ( -25.60) >DroSec_CAF1 2554 96 + 1 UUGCAUUAUGUAUCUAUCAUCUAAUCGCCUGUUGUCAAAGACAGACGAUUGCGUUGAGCAUCCAUUCAGAUACAG------AUACAUAGCUACAGCUACUAG------------------ ......(((((((((...(((((((((.(((((......))))).)))))((.....))........))))..))------)))))))((....))......------------------ ( -24.40) >DroSim_CAF1 2491 96 + 1 UUGCAUUAUGUAUCUAUCAUCUAAUCGCCUGUUGUCAAAGACAGACGAUUGCGUUGAGCAUCCAUUCAGAUACAG------AUACAUAGCUACAGCUACUAG------------------ ......(((((((((...(((((((((.(((((......))))).)))))((.....))........))))..))------)))))))((....))......------------------ ( -24.40) >DroEre_CAF1 61 105 + 1 UUGCAUUAUGUAUCUAUCAUCUAAUCGCCUGUUGUCAAAGACAGACGAUUGCGAGGAGCAUCCAUUCAGAU------ACAGCUAUGUACCAACAGCUACAGCCACAUAGCU--------- .(((((..(((((((....((((((((.(((((......))))).)))))).))((.....))....))))------)))...))))).....(((((........)))))--------- ( -28.80) >DroYak_CAF1 2557 120 + 1 UUGCAUUAUGUAUCUAUCAUCUAAUCGCCUGUUGUCAAAGACAGACGAUUGCGAGGAGCAUCCAUUCAGAUUCAGAUACAUCGACAUAGCUACAGCUACAAGCACAUAGCUGUAGCUACU .......(((((((((((.((((((((.(((((......))))).)))))).))((.....)).....)))..)))))))).....((((((((((((........)))))))))))).. ( -40.80) >consensus UUGCAUUAUGUAUCUAUCAUCUAAUCGCCUGUUGUCAAAGACAGACGAUUGCGUGGAGCAUCCAUUCAGAUACAG__ACA_AUACAUAGCUACAGCUACUAG__________________ ........(((((((.......(((((.(((((......))))).)))))((.....))........)))))))............((((....))))...................... (-18.30 = -19.30 + 1.00)


| Location | 8,490,275 – 8,490,377 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -22.94 |
| Energy contribution | -23.78 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.18 |
| Structure conservation index | 0.63 |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.999095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
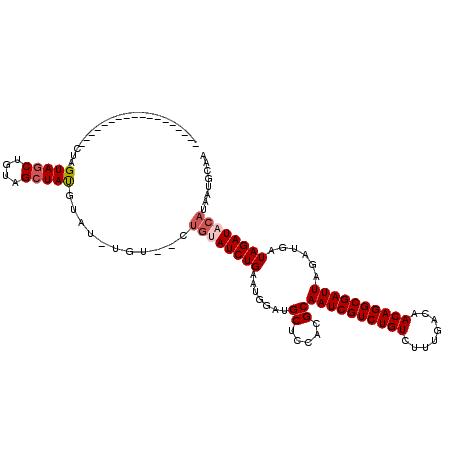
>2R_DroMel_CAF1 8490275 102 - 20766785 ------------------CUAGUAGCUGUAGCUAUGUAUUUGUAGCUGUAUCUGAAUGAAUGCUCCACGCAAUCGUCUGUCUUUGACAACAGGCGAUUAGAUGAUAGAUACAUAAUGCAG ------------------...(((((....)))))(((((((((.((((((((((.....(((.....))).((((((((........)))))))))))))).)))).)))).))))).. ( -31.60) >DroSec_CAF1 2554 96 - 1 ------------------CUAGUAGCUGUAGCUAUGUAU------CUGUAUCUGAAUGGAUGCUCAACGCAAUCGUCUGUCUUUGACAACAGGCGAUUAGAUGAUAGAUACAUAAUGCAA ------------------........((((..(((((((------((((((((((.....(((.....))).((((((((........)))))))))))))).))))))))))).)))). ( -33.40) >DroSim_CAF1 2491 96 - 1 ------------------CUAGUAGCUGUAGCUAUGUAU------CUGUAUCUGAAUGGAUGCUCAACGCAAUCGUCUGUCUUUGACAACAGGCGAUUAGAUGAUAGAUACAUAAUGCAA ------------------........((((..(((((((------((((((((((.....(((.....))).((((((((........)))))))))))))).))))))))))).)))). ( -33.40) >DroEre_CAF1 61 105 - 1 ---------AGCUAUGUGGCUGUAGCUGUUGGUACAUAGCUGU------AUCUGAAUGGAUGCUCCUCGCAAUCGUCUGUCUUUGACAACAGGCGAUUAGAUGAUAGAUACAUAAUGCAA ---------(((((((((.(..........).)))))))))((------(((((...((.....))((..((((((((((........)))))))))).))...)))))))......... ( -33.80) >DroYak_CAF1 2557 120 - 1 AGUAGCUACAGCUAUGUGCUUGUAGCUGUAGCUAUGUCGAUGUAUCUGAAUCUGAAUGGAUGCUCCUCGCAAUCGUCUGUCUUUGACAACAGGCGAUUAGAUGAUAGAUACAUAAUGCAA .((((((((((((((......))))))))))))))((..(((((((((.((((((.....(((.....))).((((((((........))))))))))))))..)))))))))...)).. ( -50.40) >consensus __________________CUAGUAGCUGUAGCUAUGUAU_UGU__CUGUAUCUGAAUGGAUGCUCCACGCAAUCGUCUGUCUUUGACAACAGGCGAUUAGAUGAUAGAUACAUAAUGCAA .....................(((((....)))))...........((((((((.......((.....))((((((((((........))))))))))......))))))))........ (-22.94 = -23.78 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:23 2006