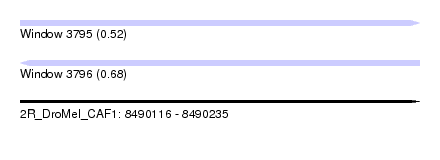
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,490,116 – 8,490,235 |
| Length | 119 |
| Max. P | 0.682995 |

| Location | 8,490,116 – 8,490,235 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.12 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -25.16 |
| Energy contribution | -26.41 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8490116 119 + 20766785 UAUCCGCAAUCGCUGCUGUCUCCAUUAAAGAUCCAUCUGGGCUUGUCAGAA-UGGCUGGCUGGUUCCGCGGAGCAGCAGCUACCACUUACUUUCAGCCAGACAGCGACGAAUCGCAAUCU .....((..(((.(((((((.(((((.(((.(((....)))))).....))-))).(((((((....(.(((((....))).)).)......)))))))))))))).)))...))..... ( -39.00) >DroSec_CAF1 2402 114 + 1 NNNNNNNNNNNNNCGCUGUUCCCAUUAAAGAUCCAUCUGGGCUGGUCAGAAAAGGCUGGCUGGUU------GGCAGCAGCUACCACUUACUUUCAGCCAGACAGCGACGAAUCGCAAUCU .............(((((((........(((....))).((((((((((......)))))((((.------(((....)))))))........))))).))))))).............. ( -30.60) >DroSim_CAF1 2338 113 + 1 UAUCAGCCAUCGCUGCUGUUCCCAUUAAAGAUCCAUCUGGGCUUGUCAGAA-UGGCUGGCUGGUU------GGCAGCAGCUACCACUUACUUUCAGCCAGACAGCGACGAAUCGCAAUCU .....((..(((.((((((..(((((.(((.(((....)))))).....))-)))((((((((.(------(((....))))..........)))))))))))))).)))...))..... ( -34.20) >DroYak_CAF1 2416 108 + 1 UAUCAGCCAUCGCUGCUGUGCCCAUUAAAGAUCCAUCUGGGCCUGUCAGAA-UGGUUGGCUGG-----------AGCAGCUACCACUUACUUUCAGCCAGACAGCGACGAAUCGCAAUCU .....((..(((.((((((..(((((..((..((....))..)).....))-)))((((((((-----------((.((......))...)))))))))))))))).)))...))..... ( -33.00) >consensus UAUCAGCCAUCGCUGCUGUUCCCAUUAAAGAUCCAUCUGGGCUUGUCAGAA_UGGCUGGCUGGUU______GGCAGCAGCUACCACUUACUUUCAGCCAGACAGCGACGAAUCGCAAUCU .........(((.((((((.............((((((((.....)))))..)))((((((((........(((....)))...........)))))))))))))).))).......... (-25.16 = -26.41 + 1.25)

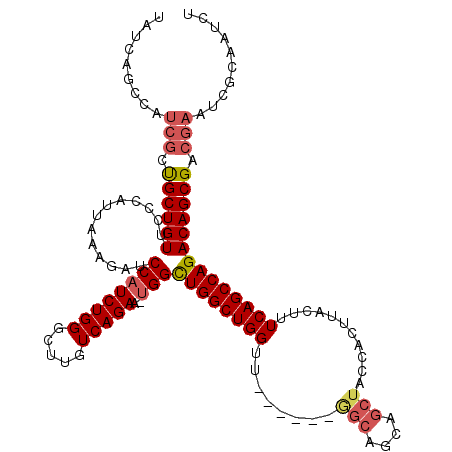
| Location | 8,490,116 – 8,490,235 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.12 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -27.08 |
| Energy contribution | -27.45 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8490116 119 - 20766785 AGAUUGCGAUUCGUCGCUGUCUGGCUGAAAGUAAGUGGUAGCUGCUGCUCCGCGGAACCAGCCAGCCA-UUCUGACAAGCCCAGAUGGAUCUUUAAUGGAGACAGCAGCGAUUGCGGAUA ...(((((((.(((.((((((((((((....(..((((.(((....)))))))..)..)))))..(((-(.(((.......)))))))...........))))))).))))))))))... ( -46.30) >DroSec_CAF1 2402 114 - 1 AGAUUGCGAUUCGUCGCUGUCUGGCUGAAAGUAAGUGGUAGCUGCUGCC------AACCAGCCAGCCUUUUCUGACCAGCCCAGAUGGAUCUUUAAUGGGAACAGCGNNNNNNNNNNNNN .....((((((((((((((((.(((((........((((((...)))))------)......)))))......)).))))...)))))))).....((....)))).............. ( -31.04) >DroSim_CAF1 2338 113 - 1 AGAUUGCGAUUCGUCGCUGUCUGGCUGAAAGUAAGUGGUAGCUGCUGCC------AACCAGCCAGCCA-UUCUGACAAGCCCAGAUGGAUCUUUAAUGGGAACAGCAGCGAUGGCUGAUA ((((.((((....)))).))))(((((........((((((...)))))------)..)))))(((((-(((((.....((((.............)))).....))).))))))).... ( -39.42) >DroYak_CAF1 2416 108 - 1 AGAUUGCGAUUCGUCGCUGUCUGGCUGAAAGUAAGUGGUAGCUGCU-----------CCAGCCAACCA-UUCUGACAGGCCCAGAUGGAUCUUUAAUGGGCACAGCAGCGAUGGCUGAUA ..........(((((((((.(((.(((..((..((((((.((((..-----------.))))..))))-))))..)))(((((.............))))).))))))))))))...... ( -38.02) >consensus AGAUUGCGAUUCGUCGCUGUCUGGCUGAAAGUAAGUGGUAGCUGCUGCC______AACCAGCCAGCCA_UUCUGACAAGCCCAGAUGGAUCUUUAAUGGGAACAGCAGCGAUGGCUGAUA ....(((((((((((((((((((((((......(((....)))((((...........)))))))))).....))).)))...)))))))).....((....)))))............. (-27.08 = -27.45 + 0.37)

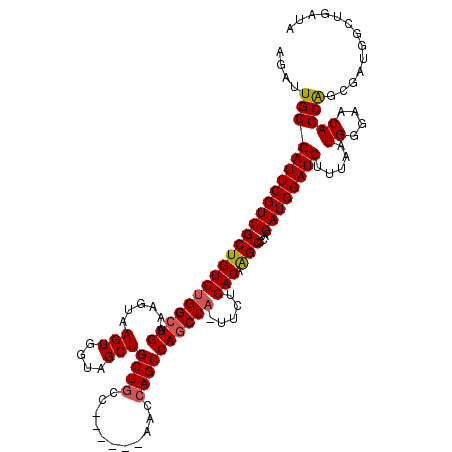
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:22 2006