
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,482,785 – 8,482,979 |
| Length | 194 |
| Max. P | 0.760262 |

| Location | 8,482,785 – 8,482,899 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.68 |
| Mean single sequence MFE | -46.08 |
| Consensus MFE | -31.05 |
| Energy contribution | -32.32 |
| Covariance contribution | 1.27 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
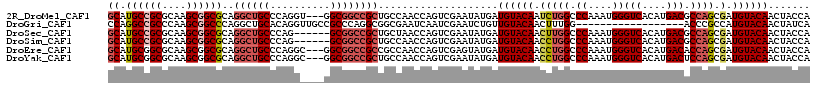
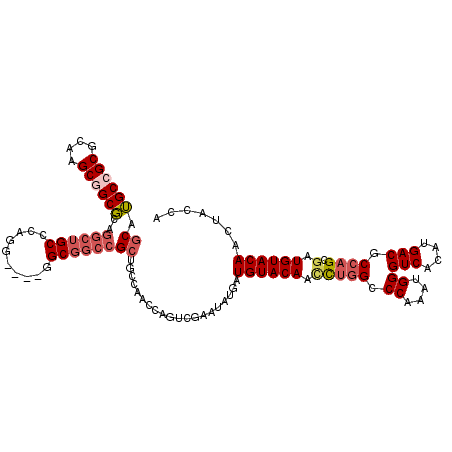
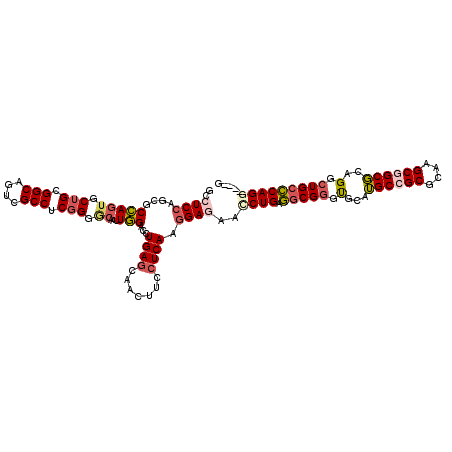
>2R_DroMel_CAF1 8482785 114 - 20766785 GCAUGCCGCGCAAGCGGCGCAGGCUGCCCAGGU---GGCGGCCGCUGCCAACCAGUCGAAUAUGAUGUACAAUCUGGCCCAAAUGGGUCACAUGACGCCAGCGAUGUACAACUACCA (((.(((((....)))))((.(((((((.....---)))))))))))).................((((((..(((((((....))(((....))))))))...))))))....... ( -49.80) >DroGri_CAF1 9952 99 - 1 CCAGGCCGCCCAAGCGGCCCAGGCUGCACAGGUUGCCGCCCAGGCGGCGAAUCAAUCGAAUCUGUUGUACAACUUUGG------------------ACCCGCCAUGUACAACUAUCA ((.((((((....))))))..)).......(((((((((....))))).))))....((....((((((((....(((------------------.....)))))))))))..)). ( -37.50) >DroSec_CAF1 7070 111 - 1 GCAUGCCGCGCAAGCGGCGCAGGCUGCCCAG------GCGGCCGCUGCUAACCAGUCGAAUAUGAUGUACAACUUGGCCCAAAUGGGUCACAUGACGCCAGCGAUGUACAACUACCA (((.(((((....)))))((.((((((....------))))))))))).................((((((.((((((((....))(((....)))))))).).))))))....... ( -47.70) >DroSim_CAF1 7095 111 - 1 GCAUGCCGCGCAAGCGGCGCAGGCUGCCCAG------GCGGCCGCUGCCAACCAGUCGAAUAUGAUGUACAACCUGGCCCAAAUGGGUCACAUGACGCCAGCGAUGUACAACUACCA (((.(((((....)))))((.((((((....------))))))))))).................((((((.((((((((....))(((....)))))))).).))))))....... ( -50.00) >DroEre_CAF1 7299 114 - 1 GCAUGCGGCGCAAGCGGCGCAGGCUGCCCAGGC---GGCGGCCGCCGCCAACCAGUCGAGUAUGAUGUACAACCUGGCCCAAAUGGGUCACAUGACACCAGCGAUGUACAACUACCA .(((((.((....))((((..(((((((.....---)))))))..))))..........))))).((((((.(((((.((....))(((....))).)))).).))))))....... ( -48.50) >DroYak_CAF1 7517 114 - 1 GCAUGCGGCGCAAGCGGCGCAGGCUGCCCAGGC---GGCGGCCGCUGCCAACCAGUCGAAUAUGAUGUACAACCUGGCCCAAAUGGGUCACAUGACUCCAGCGAUGUACAACUACCA .((((((((....))(((((.(((((((.....---))))))))).))).......))..)))).((((((.(((((.((....))(((....))).)))).).))))))....... ( -43.00) >consensus GCAUGCCGCGCAAGCGGCGCAGGCUGCCCAGG____GGCGGCCGCUGCCAACCAGUCGAAUAUGAUGUACAACCUGGCCCAAAUGGGUCACAUGACGCCAGCGAUGUACAACUACCA ((.((((((....))))))..((((((..........))))))))....................((((((.(((((.((....))(((....))).)))).).))))))....... (-31.05 = -32.32 + 1.27)
| Location | 8,482,865 – 8,482,979 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.46 |
| Mean single sequence MFE | -53.67 |
| Consensus MFE | -36.87 |
| Energy contribution | -38.07 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
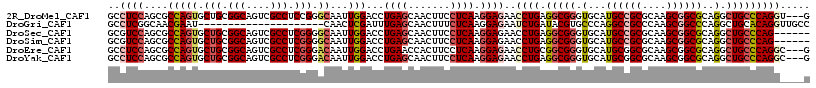
>2R_DroMel_CAF1 8482865 114 - 20766785 GCCUCCAGCGCCAGUGCUGCGGCAGUCGCCUCCGGGCAAUUGGACCUGAGCAACUUCCUCAAGGAGAACCUGAGGCGGGUGCAUGCCGCGCAAGCGGCGCAGGCUGCCCAGGU---G (((..(((((....))))).)))...(((((..(((((....(.((((.(((.((.(((((.((....))))))).)).)))..(((((....))))).))))))))))))))---) ( -57.40) >DroGri_CAF1 10014 96 - 1 GCCUCGGCAACGAAU---------------------CAACUCGAUUUGAGCAACUUUCUCAAGGAGAAUCUGAUACGUGCCCAGGCCGCCCAAGCGGCCCAGGCUGCACAGGUUGCC .....(((((((.((---------------------((..((..((((((.......))))))..))...)))).)((((((.((((((....))))))..))..))))..)))))) ( -36.70) >DroSec_CAF1 7150 111 - 1 GCGUCCAGCGCCAGUGCUGCGGCAGUCGCCUCGGGGCAAUUGGACCUGAGCAACUUCCUCAAGGAGAACCUGAGGCGGGUGCAUGCCGCGCAAGCGGCGCAGGCUGCCCAG------ (((.....)))((((.((((.(((.((((((((((((............))...((((....))))..)))))))))).)))..(((((....))))))))))))).....------ ( -56.50) >DroSim_CAF1 7175 111 - 1 GCGUCCAGCGCCAGUGCUGCGGCAGUCGCCUCGGGGCAAUUGGACCUGAGCAACUUCCUCAAGGAGAACCUGAGGCGGGUGCAUGCCGCGCAAGCGGCGCAGGCUGCCCAG------ (((.....)))((((.((((.(((.((((((((((((............))...((((....))))..)))))))))).)))..(((((....))))))))))))).....------ ( -56.50) >DroEre_CAF1 7379 114 - 1 GCCUCCAGCGCCAGUGCUGCGGCAGUCGCCUCGGGACAAUUGGACCUGAACCACUUCCUCAAGGAGAACCUGCGGCGGGUGCAUGCGGCGCAAGCGGCGCAGGCUGCCCAGGC---G ((((...(((((.(((((((((((.(((((.((((.....(((.......))).((((....))))..)))).))))).))).))))))))....))))))))).((....))---. ( -55.70) >DroYak_CAF1 7597 114 - 1 GCCUCCAGCGCCAGUGCUGCGGCAGUCGCCUCGGGACAAUUGGACCUGAGCAACUUCCUCAAGGAGAACCUGAGGCGGGUGCAUGCGGCGCAAGCGGCGCAGGCUGCCCAGGC---G ((((...(((((.(((((((((((.((((((((((.(....)..((((((.......))).)))....)))))))))).))).))))))))....))))))))).((....))---. ( -59.20) >consensus GCCUCCAGCGCCAGUGCUGCGGCAGUCGCCUCGGGGCAAUUGGACCUGAGCAACUUCCUCAAGGAGAACCUGAGGCGGGUGCAUGCCGCGCAAGCGGCGCAGGCUGCCCAGG____G ..((((....(((((.(((.(((....))).))).))...)))...((((.......)))).))))..((((.(((((.(...((((((....))))))..).)))))))))..... (-36.87 = -38.07 + 1.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:14 2006