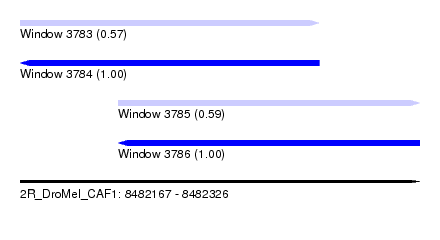
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,482,167 – 8,482,326 |
| Length | 159 |
| Max. P | 0.999465 |

| Location | 8,482,167 – 8,482,286 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -36.25 |
| Consensus MFE | -25.60 |
| Energy contribution | -26.73 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8482167 119 + 20766785 ACCG-AAGGGGGGCAGCCUAAACUCGGCUGGAUUGUGUAACAUUUUCUUUUUUUUUUUUUUUGGGUGAGGAGAAUCCACCCCCAGAUGCAGGGGUUGGAUUCCACCGUGCCACCGUAAUU .((.-...))((.(((((.......)))))((..(((...)))..))................(((..((.(((((((.((((.......)))).)))))))..))..))).))...... ( -37.90) >DroSec_CAF1 6474 103 + 1 --CGAAAGGGAGGCAUGUUUAAUGUUAAU-GUUUGUUUAACAUG-----------UUUUUUUGGGUGAGGAGAAUCCACCCCCAGGUGCAGGGGUUGGAUUC---CGUGCCACCGUAAUU --(....)((.((((((....(((((((.-......))))))).-----------................(((((((.((((.......)))).)))))))---)))))).))...... ( -35.20) >DroSim_CAF1 6498 104 + 1 --CGAAAGGGGGGCAGCUUACACUCGGCUGGAUGGUGUAACAUG-----------UUUUUUUGGGUGAGGAGAAUCCACCCCCAGGUGCAGGGGUUGGAUUC---CGUGCCACCGUAAUU --(....)((.((((.(((.(((((((..(.(((......))).-----------)....)))))))))).(((((((.((((.......)))).)))))))---..)))).))...... ( -37.10) >DroYak_CAF1 6905 97 + 1 --CGA-----GGGCAGCAUACACUCGGCUGGACUGUGUAACAU-------------UUUUUUGGGUGAGGAGAAUCCACCCCCAGGUGCAGGGGUUGGAUUC---CGUGCCACCGUAAUU --((.-----.((((.......((((.(..((..(((...)))-------------...))..).))))(.(((((((.((((.......)))).)))))))---).))))..))..... ( -34.80) >consensus __CGAAAGGGGGGCAGCUUAAACUCGGCUGGAUUGUGUAACAUG___________UUUUUUUGGGUGAGGAGAAUCCACCCCCAGGUGCAGGGGUUGGAUUC___CGUGCCACCGUAAUU ........((.((((.......((((.(..((...........................))..).))))..(((((((.((((.......)))).))))))).....)))).))...... (-25.60 = -26.73 + 1.13)

| Location | 8,482,167 – 8,482,286 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -25.58 |
| Energy contribution | -26.70 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.76 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8482167 119 - 20766785 AAUUACGGUGGCACGGUGGAAUCCAACCCCUGCAUCUGGGGGUGGAUUCUCCUCACCCAAAAAAAAAAAAAAAGAAAAUGUUACACAAUCCAGCCGAGUUUAGGCUGCCCCCCUU-CGGU ......((.(((..(((((((((((.(((((......))))))))))))....))))..................................((((.......))))))))).(..-..). ( -36.40) >DroSec_CAF1 6474 103 - 1 AAUUACGGUGGCACG---GAAUCCAACCCCUGCACCUGGGGGUGGAUUCUCCUCACCCAAAAAAA-----------CAUGUUAAACAAAC-AUUAACAUUAAACAUGCCUCCCUUUCG-- ......((.((((.(---(((((((.(((((......)))))))))))))...............-----------.(((((((......-.)))))))......)))).))......-- ( -31.90) >DroSim_CAF1 6498 104 - 1 AAUUACGGUGGCACG---GAAUCCAACCCCUGCACCUGGGGGUGGAUUCUCCUCACCCAAAAAAA-----------CAUGUUACACCAUCCAGCCGAGUGUAAGCUGCCCCCCUUUCG-- ......((..((..(---(((((((.(((((......)))))))))))))...............-----------....(((((((........).))))))))..)).........-- ( -34.00) >DroYak_CAF1 6905 97 - 1 AAUUACGGUGGCACG---GAAUCCAACCCCUGCACCUGGGGGUGGAUUCUCCUCACCCAAAAAA-------------AUGUUACACAGUCCAGCCGAGUGUAUGCUGCCC-----UCG-- ......((..(((.(---(((((((.(((((......)))))))))))))..............-------------....(((((...........))))))))..)).-----...-- ( -32.90) >consensus AAUUACGGUGGCACG___GAAUCCAACCCCUGCACCUGGGGGUGGAUUCUCCUCACCCAAAAAAA___________CAUGUUACACAAUCCAGCCGAGUGUAAGCUGCCCCCCUUUCG__ ......((((((......(((((((.(((((......))))))))))))...............................((((((...........))))))))))))........... (-25.58 = -26.70 + 1.12)

| Location | 8,482,206 – 8,482,326 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -37.92 |
| Consensus MFE | -36.24 |
| Energy contribution | -36.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
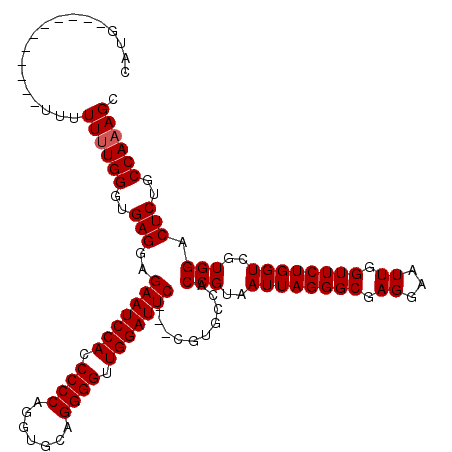
>2R_DroMel_CAF1 8482206 120 + 20766785 CAUUUUCUUUUUUUUUUUUUUUGGGUGAGGAGAAUCCACCCCCAGAUGCAGGGGUUGGAUUCCACCGUGCCACCGUAAUUAGGGCGAGGAAUUGGUUCUGGUCGUGGACUCUGCCAAAGC ..................((((((..((((.(((((((.((((.......)))).)))))))).........(((..((((((((.((...)).))))))))..))).)))..)))))). ( -37.60) >DroSec_CAF1 6511 106 + 1 CAUG-----------UUUUUUUGGGUGAGGAGAAUCCACCCCCAGGUGCAGGGGUUGGAUUC---CGUGCCACCGUAAUUAGGGCGAGGAAUUGGUUCUGGUCGUGGACUCUGCCAAAGC ....-----------...((((((..((((.(((((((.((((.......)))).)))))))---)......(((..((((((((.((...)).))))))))..))).)))..)))))). ( -37.60) >DroSim_CAF1 6536 106 + 1 CAUG-----------UUUUUUUGGGUGAGGAGAAUCCACCCCCAGGUGCAGGGGUUGGAUUC---CGUGCCACCGUAAUUAGGGCGAGGAAUUGGUUCUGGUCGUGGACUCUGCCAAAGC ....-----------...((((((..((((.(((((((.((((.......)))).)))))))---)......(((..((((((((.((...)).))))))))..))).)))..)))))). ( -37.60) >DroEre_CAF1 6731 112 + 1 CAUAU-----AUAUAUUAUUUUGGGUGAGAAGAAUCCACCCCCAGGUGCAGGGGUUGGAUUC---CGUGCCACCGUAAUUAGGGCGAGGAAUUGGUUCUGGUCGUGGACUCGGCCAAAGC .....-----........((((((.((((..(((((((.((((.......)))).)))))))---.......(((..((((((((.((...)).))))))))..))).)))).)))))). ( -40.90) >DroYak_CAF1 6938 104 + 1 CAU-------------UUUUUUGGGUGAGGAGAAUCCACCCCCAGGUGCAGGGGUUGGAUUC---CGUGCCACCGUAAUUAGGGCGAGGAAUUGGUUCUGGUCGUGGACUCUGCCAUAGC ...-------------.......(((((((.(((((((.((((.......)))).)))))))---)......(((..((((((((.((...)).))))))))..))).))).)))..... ( -35.90) >consensus CAUG___________UUUUUUUGGGUGAGGAGAAUCCACCCCCAGGUGCAGGGGUUGGAUUC___CGUGCCACCGUAAUUAGGGCGAGGAAUUGGUUCUGGUCGUGGACUCUGCCAAAGC ..................((((((..(((..(((((((.((((.......)))).)))))))..........(((..((((((((.((...)).))))))))..))).)))..)))))). (-36.24 = -36.44 + 0.20)

| Location | 8,482,206 – 8,482,326 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -30.44 |
| Energy contribution | -30.64 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8482206 120 - 20766785 GCUUUGGCAGAGUCCACGACCAGAACCAAUUCCUCGCCCUAAUUACGGUGGCACGGUGGAAUCCAACCCCUGCAUCUGGGGGUGGAUUCUCCUCACCCAAAAAAAAAAAAAAAGAAAAUG .....((.(((((((((..(((((..........((((........))))(((.((.((.......))))))).)))))..)))))))))))............................ ( -37.10) >DroSec_CAF1 6511 106 - 1 GCUUUGGCAGAGUCCACGACCAGAACCAAUUCCUCGCCCUAAUUACGGUGGCACG---GAAUCCAACCCCUGCACCUGGGGGUGGAUUCUCCUCACCCAAAAAAA-----------CAUG .....((.(((((((((..((((...........((((........))))(((.(---(........)).)))..))))..))))))))))).............-----------.... ( -34.00) >DroSim_CAF1 6536 106 - 1 GCUUUGGCAGAGUCCACGACCAGAACCAAUUCCUCGCCCUAAUUACGGUGGCACG---GAAUCCAACCCCUGCACCUGGGGGUGGAUUCUCCUCACCCAAAAAAA-----------CAUG .....((.(((((((((..((((...........((((........))))(((.(---(........)).)))..))))..))))))))))).............-----------.... ( -34.00) >DroEre_CAF1 6731 112 - 1 GCUUUGGCCGAGUCCACGACCAGAACCAAUUCCUCGCCCUAAUUACGGUGGCACG---GAAUCCAACCCCUGCACCUGGGGGUGGAUUCUUCUCACCCAAAAUAAUAUAU-----AUAUG ..(((((.((......)).)))))......................(((((...(---(((((((.(((((......)))))))))))))..))))).............-----..... ( -31.90) >DroYak_CAF1 6938 104 - 1 GCUAUGGCAGAGUCCACGACCAGAACCAAUUCCUCGCCCUAAUUACGGUGGCACG---GAAUCCAACCCCUGCACCUGGGGGUGGAUUCUCCUCACCCAAAAAA-------------AUG .....((.(((((((((..((((...........((((........))))(((.(---(........)).)))..))))..)))))))))))............-------------... ( -34.00) >consensus GCUUUGGCAGAGUCCACGACCAGAACCAAUUCCUCGCCCUAAUUACGGUGGCACG___GAAUCCAACCCCUGCACCUGGGGGUGGAUUCUCCUCACCCAAAAAAA___________CAUG ..(((((..(((.......................(((((......)).)))......(((((((.(((((......))))))))))))..)))..)))))................... (-30.44 = -30.64 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:12 2006