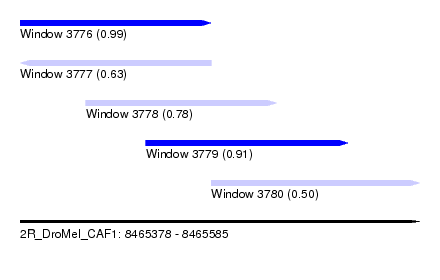
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,465,378 – 8,465,585 |
| Length | 207 |
| Max. P | 0.988384 |

| Location | 8,465,378 – 8,465,477 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -26.26 |
| Energy contribution | -26.97 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8465378 99 + 20766785 GUGUGUAGGCCGGAGGCCUGUCA------GGAAAUGGCAUCGCAGCCAAAUCCGGAUAUGGAUAUGGUUUGGCCGGCAGUGGCGGCGCCAUUGCCGGUGGAGAAG .((((.(((((...)))))((((------.....))))..))))((((.(((((....))))).))))((.((((((((((((...)))))))))))).)).... ( -46.60) >DroSec_CAF1 8945 99 + 1 GUGUGUAGGCCGGAGGCCUGUCA------GGAAAUGGCAUGGCAGCCAAAUCCGGAUAGGGAUAUGGUUUGGCCGGCAGUGGCGGCGCCAUGGCCAGUGGCGAAG ........((((..((((.((((------.....))))((((((((((.((((......)))).)))))..((((.(...).)))))))))))))..)))).... ( -43.10) >DroEre_CAF1 10418 99 + 1 GUGUGUAAGCCGGAGGCCUGUCA------GGAAAUGGCAUGGCAGCCAAAUUCGGAUAGGGAUAUGGCUUGGCUGACAGUGGCGGCGCCAUUGCCGGUGGAGAAG .....(((((((....((((((.------(((..((((......))))..))).))))))....)))))))((((.(((((((...))))))).))))....... ( -38.80) >DroYak_CAF1 9533 99 + 1 GUGUGUAAGCCGGAGGCCUGUCA------GGAAAUGGCAUAGCAGCCAAGUGCGGAUAGGGGUAUGGUUUGGCUGGCAGUGGCGGCGCCAUUGCCGGUGGAGAUG .....(((((((....((((((.------(.(..((((......))))..).).))))))....)))))))((((((((((((...))))))))))))....... ( -43.00) >DroAna_CAF1 9739 105 + 1 GCGUAUAAGCCGGCGGUCUGUCCUUGAUUGGAAAAGGCAUAGCAGCUAGGUCCGGAUAGGGAUAGGGCUUGAGGGCCACCGGAGGAGCGGGUGGUGGUGGCGAGG ........((((.((.((((((((((.(((((...(((......)))...))))).)))))))))).)......((((((.(.....).))))))).)))).... ( -42.30) >DroPer_CAF1 8997 105 + 1 GCGUAUAAGCUGGUGGCCUGUCGGUCAUUGGGAACGGCAUUGCGGCCAGAUGCGGAUACUUAUACGGCUUCUCAACCACAGGCGGAGCGGGUGUAGGUGGAGACG .(((((...(((((.((.(((((.((.....)).)))))..)).))))))))))...((((((((.(((((.(........).)))))..))))))))(....). ( -36.40) >consensus GUGUGUAAGCCGGAGGCCUGUCA______GGAAAUGGCAUAGCAGCCAAAUCCGGAUAGGGAUAUGGCUUGGCCGGCAGUGGCGGCGCCAUUGCCGGUGGAGAAG .....(((((((....((((((.......(((..((((......))))..))).))))))....)))))))...(((((((((...))))))))).......... (-26.26 = -26.97 + 0.70)


| Location | 8,465,378 – 8,465,477 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -13.41 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8465378 99 - 20766785 CUUCUCCACCGGCAAUGGCGCCGCCACUGCCGGCCAAACCAUAUCCAUAUCCGGAUUUGGCUGCGAUGCCAUUUCC------UGACAGGCCUCCGGCCUACACAC ........((((((.((((...)))).))))))................((.(((..((((......))))..)))------.)).(((((...)))))...... ( -36.50) >DroSec_CAF1 8945 99 - 1 CUUCGCCACUGGCCAUGGCGCCGCCACUGCCGGCCAAACCAUAUCCCUAUCCGGAUUUGGCUGCCAUGCCAUUUCC------UGACAGGCCUCCGGCCUACACAC ....(((.(((((..((((...))))..)))))............(((.((.(((..((((......))))..)))------.)).))).....)))........ ( -34.50) >DroEre_CAF1 10418 99 - 1 CUUCUCCACCGGCAAUGGCGCCGCCACUGUCAGCCAAGCCAUAUCCCUAUCCGAAUUUGGCUGCCAUGCCAUUUCC------UGACAGGCCUCCGGCUUACACAC ..........((((.((((...)))).))))....(((((.....(((.((.(((..((((......)))).))).------.)).))).....)))))...... ( -27.10) >DroYak_CAF1 9533 99 - 1 CAUCUCCACCGGCAAUGGCGCCGCCACUGCCAGCCAAACCAUACCCCUAUCCGCACUUGGCUGCUAUGCCAUUUCC------UGACAGGCCUCCGGCUUACACAC ..........((((.((((...)))).))))((((..........(((.((.(....((((......))))....)------.)).))).....))))....... ( -24.26) >DroAna_CAF1 9739 105 - 1 CCUCGCCACCACCACCCGCUCCUCCGGUGGCCCUCAAGCCCUAUCCCUAUCCGGACCUAGCUGCUAUGCCUUUUCCAAUCAAGGACAGACCGCCGGCUUAUACGC ....((((((...............))))))....(((((...(((......)))....((.(((...((((........))))..)).).)).)))))...... ( -22.76) >DroPer_CAF1 8997 105 - 1 CGUCUCCACCUACACCCGCUCCGCCUGUGGUUGAGAAGCCGUAUAAGUAUCCGCAUCUGGCCGCAAUGCCGUUCCCAAUGACCGACAGGCCACCAGCUUAUACGC ..((((.(((.(((..((...))..)))))).))))...(((((((((..........(((......)))........((.((....)).))...))))))))). ( -25.60) >consensus CUUCUCCACCGGCAAUGGCGCCGCCACUGCCAGCCAAACCAUAUCCCUAUCCGGAUUUGGCUGCCAUGCCAUUUCC______UGACAGGCCUCCGGCUUACACAC ..........((((.((((...)))).))))..........................((((......))))...............((((.....))))...... (-13.41 = -14.38 + 0.97)


| Location | 8,465,412 – 8,465,511 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.62 |
| Mean single sequence MFE | -41.95 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8465412 99 + 20766785 CGCAGCCAAAUCC---GGAUAUGGAUAUGGUUUGGCCGGCAGUGGCGGC---GCCAUUGCCGGUGGAGAAGGUGGUGGCGGCGGCGAUAGUGGACACUUUUCCGG ((.(((((.((((---(....))))).)))))))((((((((((((...---))))))))))))(((((((((.((.((....)).)).....)).))))))).. ( -44.70) >DroSec_CAF1 8979 99 + 1 GGCAGCCAAAUCC---GGAUAGGGAUAUGGUUUGGCCGGCAGUGGCGGC---GCCAUGGCCAGUGGCGAAGGUGGUGGCGGCGGCGUUAGUGGACACUUUUCCGG (((.((((.((((---......)))).))))...)))..((.(((((.(---(((...((((.(.((....)).))))))))).))))).))............. ( -41.50) >DroEre_CAF1 10452 99 + 1 GGCAGCCAAAUUC---GGAUAGGGAUAUGGCUUGGCUGACAGUGGCGGC---GCCAUUGCCGGUGGAGAAGGUGGUGGCGGCGGCGAGAGCGGACACUUUUCCGG ((((((((.((((---......)))).)))))..)))...((((.((.(---(((((..((.........))..)))))).))((....))...))))....... ( -40.50) >DroYak_CAF1 9567 99 + 1 AGCAGCCAAGUGC---GGAUAGGGGUAUGGUUUGGCUGGCAGUGGCGGC---GCCAUUGCCGGUGGAGAUGGGGGUGGCGGCGGUGUGAGCGGACACUUUUCCGG .((.((((.((((---........))))..(((.((((((((((((...---)))))))))))).))).......)))).))........((((......)))). ( -38.10) >DroMoj_CAF1 14050 105 + 1 CGUAUCAAAGUCCGGCGGAUAGGGAUACGGCUUGCGUGCCACUGGCGGCUUAGCCAGCACCGCUGGUGGCGGUGGCGGUGGCGGUGUCGGUGGACACUUCUCGGC ((((((...((((...))))...))))))......((((((((((((.(...((((.(.((((((....)))))).).))))).))))))))).)))........ ( -46.90) >DroAna_CAF1 9779 99 + 1 AGCAGCUAGGUCC---GGAUAGGGAUAGGGCUUGAGGGCCACCGGAGGA---GCGGGUGGUGGUGGCGAGGGAGGUGGCGGCGGGGUGAGUGGGCACUUCUCCGG .((((((..((((---......))))..)))).....((((((.(....---.).))))))....))(.((((((((.(.((.......)).).))))))))).. ( -40.00) >consensus AGCAGCCAAAUCC___GGAUAGGGAUAUGGCUUGGCUGGCAGUGGCGGC___GCCAUUGCCGGUGGAGAAGGUGGUGGCGGCGGCGUGAGUGGACACUUUUCCGG ...(((((.((((.........)))).)))))..((((((((((((......))))))))))))......((.((((.(.((.......)).).))))...)).. (-27.33 = -27.75 + 0.42)

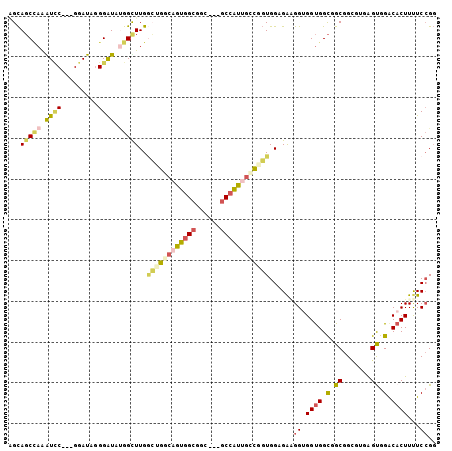
| Location | 8,465,443 – 8,465,548 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.80 |
| Mean single sequence MFE | -52.13 |
| Consensus MFE | -31.53 |
| Energy contribution | -32.53 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8465443 105 + 20766785 GCCGGCAGUGGCGGC---GCCAUUGCCGGUGGAGAAGGUGGUGGCGGCGGCGAUAGUGGACACUUUUCCGGCUCCUCUAGUGGAGGAGGUGGUGGCGGCAGCUGCCAG ((((((((((((...---))))))))))))......((..((.((.((.((.((..((((......)))).((((((.....)))))))).)).)).)).))..)).. ( -55.80) >DroSec_CAF1 9010 105 + 1 GCCGGCAGUGGCGGC---GCCAUGGCCAGUGGCGAAGGUGGUGGCGGCGGCGUUAGUGGACACUUUUCCGGCUCCUCUAGUGGAGGAGGUGGUGGCGGCAGCUGCCAG ((((.((.(((((.(---(((...((((.(.((....)).))))))))).))))).)).........(((.((((((.....)))))).)))))))(((....))).. ( -51.70) >DroEre_CAF1 10483 105 + 1 GCUGACAGUGGCGGC---GCCAUUGCCGGUGGAGAAGGUGGUGGCGGCGGCGAGAGCGGACACUUUUCCGGCUGCUCUAGUGGAGGAGGUGGUGGUGGCAGCUGCCAG ........(((((((---(((((..((.........))..))))).((.((....))...((((...(((.((.(((.....))).)).))).)))))).))))))). ( -50.30) >DroYak_CAF1 9598 102 + 1 GCUGGCAGUGGCGGC---GCCAUUGCCGGUGGAGAUGGGGGUGGCGGCGGUGUGAGCGGACACUUUUCCGGCUCCUCCAGUGGAGGAGGUG---GUGGCAGUUGCCAG ((((((((((((...---)))))))))))).........((..((.(((((((......)))))...(((.(((((((...))))))).))---)..)).))..)).. ( -51.70) >DroMoj_CAF1 14084 108 + 1 CGUGCCACUGGCGGCUUAGCCAGCACCGCUGGUGGCGGUGGCGGUGGCGGUGUCGGUGGACACUUCUCGGCCAACUCCAAUGCAUCCGGUGGUGGGGGCAGCUGCCAG .......(((((((((..(((..(((((((((..(((.(((.(.((((((((((....)))))).....)))).).))).)))..)))))))))..)))))))))))) ( -59.90) >DroPer_CAF1 9068 102 + 1 UCAACCACAGGCGGA---GCGGGUGUAGGUGGAGACGGGGGCGGCGGCGGUGUUAGUGGACACUUCUCCAGCUCCUCCGGCGGCGG---CGGCGGUGGCAGCUGCCAG .........(.((((---(.((((......(....)((((((....))((((((....)))))).)))).))))))))).)(((((---(.((....)).)))))).. ( -43.40) >consensus GCUGGCAGUGGCGGC___GCCAUUGCCGGUGGAGAAGGUGGUGGCGGCGGCGUUAGUGGACACUUUUCCGGCUCCUCCAGUGGAGGAGGUGGUGGUGGCAGCUGCCAG ((((.(((((((......))))))).)))).........((((((.((.((.....((((......)))).(((((((...)))))))......)).)).)))))).. (-31.53 = -32.53 + 1.00)

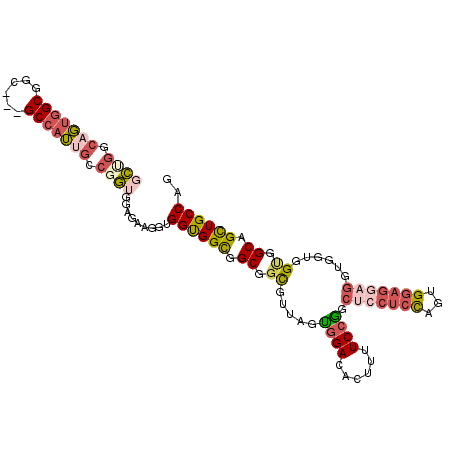
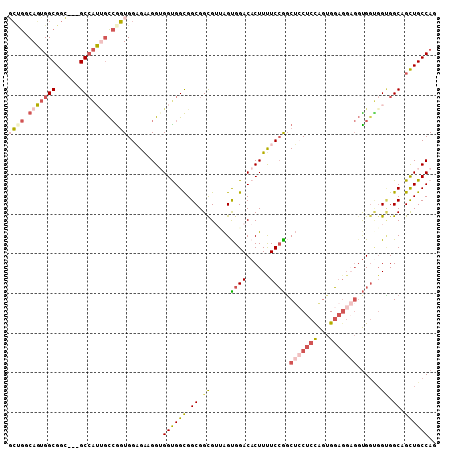
| Location | 8,465,477 – 8,465,585 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -49.12 |
| Consensus MFE | -32.03 |
| Energy contribution | -34.95 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8465477 108 + 20766785 GUGGUGGCGGCGGCGAUAGUGGACACUUUUCCGGCUCCUCUAGUGGAGGAGGUGGUGGCGGCAGCUGCCAGCCAGGAACCACUCUGCGCUCCUCCACCAAAAUCUCAU .((((((.((.((((..(((((........(((.((((((.....)))))).)))(((((((....))).))))....)))))...)))))).))))))......... ( -52.90) >DroSec_CAF1 9044 108 + 1 GUGGUGGCGGCGGCGUUAGUGGACACUUUUCCGGCUCCUCUAGUGGAGGAGGUGGUGGCGGCAGCUGCCAGCCAGGAACCACUCUGCGCUCCUCCACCAAAAUUUCGU .((((((.((.(((((.(((((........(((.((((((.....)))))).)))(((((((....))).))))....)))))..))))))).))))))......... ( -54.50) >DroEre_CAF1 10517 108 + 1 GUGGUGGCGGCGGCGAGAGCGGACACUUUUCCGGCUGCUCUAGUGGAGGAGGUGGUGGUGGCAGCUGCCAGCCAGGUACCACUCGACGCUCCUCCACCAAUAUCUCGU .((((((.((.(((.(((((((.(........).))))))).((.(((..((((.(((((((....)))).)))..)))).))).))))))).))))))......... ( -47.40) >DroYak_CAF1 9632 105 + 1 GGGGUGGCGGCGGUGUGAGCGGACACUUUUCCGGCUCCUCCAGUGGAGGAGGUG---GUGGCAGUUGCCAGCCGGGCACCACUCUGCGCUCCUCCACCAAAAUCUCGU ..(((((.((.((((((((.((..((((..(((.(((((((...))))))).))---)..).)))((((.....)))))).))).))))))).))))).......... ( -46.90) >DroAna_CAF1 9844 105 + 1 GAGGUGGCGGCGGGGUGAGUGGGCACUUCUCCGGCUCCUCUAGAGGCG---GCGGCGGUGGCAGACUCCAUCCGGCAACCACACGUCGCUCCUCCACCAGAAUCUCAU ((((.(.(((.((((((......)))))).))).).))))..((((.(---((((((((((..(.((......)))..)))).))))))))))).............. ( -44.60) >DroPer_CAF1 9102 105 + 1 GGGGCGGCGGCGGUGUUAGUGGACACUUCUCCAGCUCCUCCGGCGGCGG---CGGCGGUGGCAGCUGCCAGCCGGGCACAACCCUCCUCUCCUCCACGAGGAUCUCGU (((((((.((.((((((....)))))).)))).)))))(((((((((((---(.((....)).)))))).))))))........(((((........)))))...... ( -48.40) >consensus GUGGUGGCGGCGGCGUGAGUGGACACUUUUCCGGCUCCUCUAGUGGAGGAGGUGGUGGUGGCAGCUGCCAGCCAGGAACCACUCUGCGCUCCUCCACCAAAAUCUCGU ..(((((.((.((((..(((((........(((.(((((((...))))))).)))(((((((....))).))))....)))))...)))))).))))).......... (-32.03 = -34.95 + 2.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:06 2006