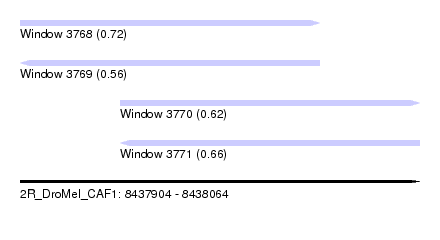
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,437,904 – 8,438,064 |
| Length | 160 |
| Max. P | 0.721214 |

| Location | 8,437,904 – 8,438,024 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -41.65 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.35 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8437904 120 + 20766785 AGAAGGCGCUGAAGGAUGUGUGCAUCAAUCUGCUGAUGGAGGUCUUGGACCGUGUAAACGAUGUCUUCUUGGAGCGUGCUGUGCGCCUUCUGCAGACAUUGCGCAAGCAAUCCAAUAUCA (((((((((...((.((((...(((((......)))))....(((.(((((((....)))..))))....))))))).))..)))))))))......(((((....)))))......... ( -40.60) >DroVir_CAF1 7700 120 + 1 AGAAAGCGUUAAAGGAUGUCUGCCUCAAUCUGCUGAUGGAGGUGCUGGACCGUGUGAAUGAUGUAUACCUGGAGCGUAUCGUUCGCUUCCUGCAGACAUUGCGCAAACUGUCCAGCAUUA .....(((((....)))))..(((((.(((....))).)))))(((((((.((.((..((((((......((((((.......))))))......))))))..)).)).))))))).... ( -40.30) >DroGri_CAF1 1147 120 + 1 AGAAAUCGCUUAAGGAUGUAUGCCUCAAUCUGCUGAUGGAGGUGCUGGAUCGUGUGAACGAUGUUUACCUGGAGCGUAUCGUUCGCUUCCUGCAGGCAUUGCGCAAGCAAUCCGUCAUUA ..............(((....(((((.(((....))).)))))((.(((....(((((((((((((.....))))..))))))))).))).)).((.(((((....)))))))))).... ( -40.60) >DroWil_CAF1 1146 120 + 1 AGAAAUCACUGAAGGAUAUCUGCAUCAAUUUGCUAAUGGAAGUGUUGGACCGUGUGAACGAUGUCUACCUGGAACGCGUCGUGCGUUUCUUGCAGACAUUGCGCAAACAGUCCAACAUUA (((.(((.(....)))).)))(((......))).......((((((((((.((.((..((((((((.(..(((((((.....)))))))..).))))))))..)).)).)))))))))). ( -42.70) >DroMoj_CAF1 1094 120 + 1 AAAAAUCGCUCAAGGAUGUCUGUCUCAAUCUAUUGAUGGAGGUGCUUGACCGUGUCAACGACGUCUAUUUGGAACGUGUCGUGCGCUUCUUGCAGACAUUGCGCAAGCAGGCCAACAUCA .......(((....(((((((((....(((....)))((((((((.(((((((....)))((((((....)).)))))))).)))))))).)))))))))((....)).)))........ ( -38.80) >DroAna_CAF1 1097 120 + 1 AGAAGUCGCUGAAGGAUGUGUGCAUCAGCCUGUUGAUGGAGGUGUUGGACCGUGUGAACGAUGUCUACCUGGAGCGUGUCGUGCGCUUCCUACAGACAUUGCGCAAACAGUCCAACAUCA ......(((........)))..((((((....))))))..((((((((((.((.((..((((((((....(((((((.....)))))))....))))))))..)).)).)))))))))). ( -46.90) >consensus AGAAAUCGCUGAAGGAUGUCUGCAUCAAUCUGCUGAUGGAGGUGCUGGACCGUGUGAACGAUGUCUACCUGGAGCGUGUCGUGCGCUUCCUGCAGACAUUGCGCAAACAGUCCAACAUCA ........................((.(((....))).))((((((((((.((.((..((((((((.(..(((((((.....)))))))..).))))))))..)).)).)))))))))). (-29.20 = -29.35 + 0.15)


| Location | 8,437,904 – 8,438,024 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -24.08 |
| Energy contribution | -24.67 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8437904 120 - 20766785 UGAUAUUGGAUUGCUUGCGCAAUGUCUGCAGAAGGCGCACAGCACGCUCCAAGAAGACAUCGUUUACACGGUCCAAGACCUCCAUCAGCAGAUUGAUGCACACAUCCUUCAGCGCCUUCU .....(((((.(((((((((..((....))....))))).))))...)))))((((.(...........((((...)))).......((.((..((((....))))..)).))).)))). ( -32.60) >DroVir_CAF1 7700 120 - 1 UAAUGCUGGACAGUUUGCGCAAUGUCUGCAGGAAGCGAACGAUACGCUCCAGGUAUACAUCAUUCACACGGUCCAGCACCUCCAUCAGCAGAUUGAGGCAGACAUCCUUUAACGCUUUCU ...((((((((.((.((....((((.(((.(((.(((.......))))))..))).))))....)).)).))))))))((((.(((....))).))))...................... ( -34.70) >DroGri_CAF1 1147 120 - 1 UAAUGACGGAUUGCUUGCGCAAUGCCUGCAGGAAGCGAACGAUACGCUCCAGGUAAACAUCGUUCACACGAUCCAGCACCUCCAUCAGCAGAUUGAGGCAUACAUCCUUAAGCGAUUUCU .......((((((((((.((..(((((...(((.(((.......)))))))))))...(((((....)))))...)).((((.(((....))).))))..........)))))))))).. ( -31.50) >DroWil_CAF1 1146 120 - 1 UAAUGUUGGACUGUUUGCGCAAUGUCUGCAAGAAACGCACGACGCGUUCCAGGUAGACAUCGUUCACACGGUCCAACACUUCCAUUAGCAAAUUGAUGCAGAUAUCCUUCAGUGAUUUCU ...(((((((((((.((.((.((((((((..(.(((((.....))))).)..)))))))).)).)).))))))))))).........(((......)))(((.((((....).))).))) ( -42.90) >DroMoj_CAF1 1094 120 - 1 UGAUGUUGGCCUGCUUGCGCAAUGUCUGCAAGAAGCGCACGACACGUUCCAAAUAGACGUCGUUGACACGGUCAAGCACCUCCAUCAAUAGAUUGAGACAGACAUCCUUGAGCGAUUUUU ...(((((((((.((((((.......)))))).)).)).)))))(((((......((.(((.(((((...)))))....(((.(((....))).)))...))).))...)))))...... ( -30.30) >DroAna_CAF1 1097 120 - 1 UGAUGUUGGACUGUUUGCGCAAUGUCUGUAGGAAGCGCACGACACGCUCCAGGUAGACAUCGUUCACACGGUCCAACACCUCCAUCAACAGGCUGAUGCACACAUCCUUCAGCGACUUCU ...(((((((((((.((.((.(((((((..(((.(((.......))))))...))))))).)).)).))))))))))).............(((((............)))))....... ( -40.90) >consensus UAAUGUUGGACUGCUUGCGCAAUGUCUGCAGGAAGCGCACGACACGCUCCAGGUAGACAUCGUUCACACGGUCCAACACCUCCAUCAGCAGAUUGAGGCAGACAUCCUUCAGCGAUUUCU ...((((((((((..((.((.((((((((..(.((((.......)))).)..)))))))).)).))..))))))))))..((.(((....))).))........................ (-24.08 = -24.67 + 0.59)


| Location | 8,437,944 – 8,438,064 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -40.17 |
| Consensus MFE | -28.04 |
| Energy contribution | -28.43 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8437944 120 + 20766785 GGUCUUGGACCGUGUAAACGAUGUCUUCUUGGAGCGUGCUGUGCGCCUUCUGCAGACAUUGCGCAAGCAAUCCAAUAUCAAGCCGAACAUCGUGGUCGAGAUUUUCAAGCAGGUGGUCAA (((.(((((.(((....))).(((((.(..((((.((((...)))))))).).)))))((((....))))))))).))).....((.((((.((.(.(((...))).).)))))).)).. ( -33.40) >DroVir_CAF1 7740 120 + 1 GGUGCUGGACCGUGUGAAUGAUGUAUACCUGGAGCGUAUCGUUCGCUUCCUGCAGACAUUGCGCAAACUGUCCAGCAUUAAGCCGAAUGUCAUCGUUGAGAUCUUCAAGCAACUGGUGAA ((((((((((.((.((..((((((......((((((.......))))))......))))))..)).)).))))))))))..........(((((((((.(.....)...)))).))))). ( -38.40) >DroGri_CAF1 1187 120 + 1 GGUGCUGGAUCGUGUGAACGAUGUUUACCUGGAGCGUAUCGUUCGCUUCCUGCAGGCAUUGCGCAAGCAAUCCGUCAUUAAGCCGAAUGUGAUCGUUGAGAUCUUCAAGCAGUUGGUCAA ..(((.(((....(((((((((((((.....))))..))))))))).))).)))((.(((((....)))))))........(((((.(((((((.....)))).....))).)))))... ( -40.30) >DroWil_CAF1 1186 120 + 1 AGUGUUGGACCGUGUGAACGAUGUCUACCUGGAACGCGUCGUGCGUUUCUUGCAGACAUUGCGCAAACAGUCCAACAUUAAACCCAAUAUCCUUGUCGAAGUCUUCAAGCAGCUGGUGAA ((((((((((.((.((..((((((((.(..(((((((.....)))))))..).))))))))..)).)).))))))))))...((((.....((((..(....)..))))....))).).. ( -42.00) >DroMoj_CAF1 1134 120 + 1 GGUGCUUGACCGUGUCAACGACGUCUAUUUGGAACGUGUCGUGCGCUUCUUGCAGACAUUGCGCAAGCAGGCCAACAUCAAGCCAAAUAUCAUCGUUGAGAUCUUCAAGCAGUUGGUCAA ..(((((((..((.(((((((.(..(((((((...((((.(.((((.....)).....((((....))))))).))))....))))))).).))))))).))..)))))))......... ( -41.60) >DroAna_CAF1 1137 120 + 1 GGUGUUGGACCGUGUGAACGAUGUCUACCUGGAGCGUGUCGUGCGCUUCCUACAGACAUUGCGCAAACAGUCCAACAUCAAGCCGAACGUGAUCGUCGAGGUAUUCAAGCAGCUCGUCAA ((((((((((.((.((..((((((((....(((((((.....)))))))....))))))))..)).)).))))))))))...............(.((((.(........).)))).).. ( -45.30) >consensus GGUGCUGGACCGUGUGAACGAUGUCUACCUGGAGCGUGUCGUGCGCUUCCUGCAGACAUUGCGCAAACAGUCCAACAUCAAGCCGAAUAUCAUCGUCGAGAUCUUCAAGCAGCUGGUCAA ((((((((((.((.((..((((((((.(..(((((((.....)))))))..).))))))))..)).)).))))))))))......................................... (-28.04 = -28.43 + 0.39)

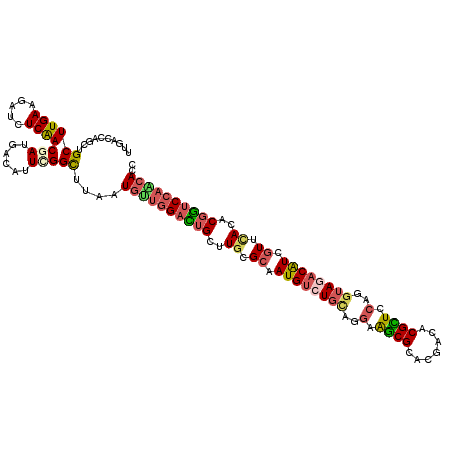
| Location | 8,437,944 – 8,438,064 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -24.72 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8437944 120 - 20766785 UUGACCACCUGCUUGAAAAUCUCGACCACGAUGUUCGGCUUGAUAUUGGAUUGCUUGCGCAAUGUCUGCAGAAGGCGCACAGCACGCUCCAAGAAGACAUCGUUUACACGGUCCAAGACC ..((((....(.((((.....)))).)((((((((...((((.....((..(((((((((..((....))....))))).))))..)).))))..))))))))......))))....... ( -32.70) >DroVir_CAF1 7740 120 - 1 UUCACCAGUUGCUUGAAGAUCUCAACGAUGACAUUCGGCUUAAUGCUGGACAGUUUGCGCAAUGUCUGCAGGAAGCGAACGAUACGCUCCAGGUAUACAUCAUUCACACGGUCCAGCACC ..........(((.(((..((........))..))))))....((((((((.((.((....((((.(((.(((.(((.......))))))..))).))))....)).)).)))))))).. ( -32.80) >DroGri_CAF1 1187 120 - 1 UUGACCAACUGCUUGAAGAUCUCAACGAUCACAUUCGGCUUAAUGACGGAUUGCUUGCGCAAUGCCUGCAGGAAGCGAACGAUACGCUCCAGGUAAACAUCGUUCACACGAUCCAGCACC .........((((.((.((((.....))))....(((((((..((..(((((((....))))).))..))..))))(((((((..((.....))....)))))))...))))).)))).. ( -33.00) >DroWil_CAF1 1186 120 - 1 UUCACCAGCUGCUUGAAGACUUCGACAAGGAUAUUGGGUUUAAUGUUGGACUGUUUGCGCAAUGUCUGCAAGAAACGCACGACGCGUUCCAGGUAGACAUCGUUCACACGGUCCAACACU ....((((.(.((((..(....)..)))).)..))))......(((((((((((.((.((.((((((((..(.(((((.....))))).)..)))))))).)).)).))))))))))).. ( -45.30) >DroMoj_CAF1 1134 120 - 1 UUGACCAACUGCUUGAAGAUCUCAACGAUGAUAUUUGGCUUGAUGUUGGCCUGCUUGCGCAAUGUCUGCAAGAAGCGCACGACACGUUCCAAAUAGACGUCGUUGACACGGUCAAGCACC .........(((((((.....(((((((((.(((((((..((.(((((((((.((((((.......)))))).)).)).)))))))..)))))))..))))))))).....))))))).. ( -46.90) >DroAna_CAF1 1137 120 - 1 UUGACGAGCUGCUUGAAUACCUCGACGAUCACGUUCGGCUUGAUGUUGGACUGUUUGCGCAAUGUCUGUAGGAAGCGCACGACACGCUCCAGGUAGACAUCGUUCACACGGUCCAACACC ....(((((((...((.....))((((....))))))))))).(((((((((((.((.((.(((((((..(((.(((.......))))))...))))))).)).)).))))))))))).. ( -44.30) >consensus UUGACCAGCUGCUUGAAGAUCUCAACGAUGACAUUCGGCUUAAUGUUGGACUGCUUGCGCAAUGUCUGCAGGAAGCGCACGACACGCUCCAGGUAGACAUCGUUCACACGGUCCAACACC ..........((((((.....))))(((......)))))....((((((((((..((.((.((((((((..(.((((.......)))).)..)))))))).)).))..)))))))))).. (-24.72 = -25.42 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:58 2006