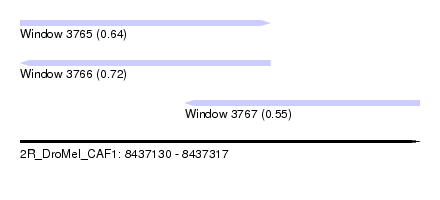
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,437,130 – 8,437,317 |
| Length | 187 |
| Max. P | 0.720347 |

| Location | 8,437,130 – 8,437,247 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -42.98 |
| Consensus MFE | -27.81 |
| Energy contribution | -29.45 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
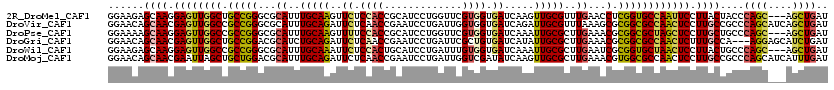
>2R_DroMel_CAF1 8437130 117 + 20766785 AUCAGCU---GCUGGGUAGUAAGGAAUUGGCACCGAGGUUCAAACGCAACUUGAUCACCACGAACCAGGAUGCGGUGGAGAACUUGCAAAUGCGCCCGGCAGCCAACUCCUUGCUCUUCC .......---...(((.((((((((.(((((.(((.(((.((...((((.((..(((((.((........)).)))))..)).))))...)).))))))..))))).))))))))..))) ( -40.10) >DroVir_CAF1 6947 120 + 1 AUCAGCUGAUGCUGGGCGGCAAGGAGUUGGCGCCGCGCUUUAAACGCAAUCUGAUCACCACCAAUCAGGAUUCGGUUGAGAAUCUGCAAAUGCGCCCGGCGGCCAACUCGUUGCUGUUCC .(((((....)))))(((((((.(((((((((((((((.......(((..(((((........)))))(((((......))))))))....))))..))).)))))))).)))))))... ( -54.00) >DroPse_CAF1 25583 117 + 1 AUCAGCU---GCUGGGCAGCAAGGAGCUAGCGCCGCGUUUCAAGCGCAAUUUGAUCACCACGAACCAGGAUGCGGUGGAAAACUUGCAAAUGCGCCCGGCGGCCAACUCCUUGCUUUUCC ....(((---(((((((.(((((...(((.(((.((((.....))))..........((........))..))).)))....)))))......))))))))))................. ( -40.30) >DroGri_CAF1 313 117 + 1 AUCAGAUGCUCCU---UGGCAAAGAGUUGGCGCCGCGUUUCAAGCGCAAUAUGAUCACAGCGAAUCAGGAUUCGGUUGAGAAUCUGCAGAUGCGUCCGGCAGCCAACUCGUUGCUGUUCC .............---.(((((.(((((((((((((((.....))))............(((.(((.((((((......))))))...))).)))..))).)))))))).)))))..... ( -42.80) >DroWil_CAF1 267 117 + 1 AUCAGCU---GCUGGGCAGUAAGGAGUUAGCACCGCGAUUCAAGCGCAAUUUGAUCACCACAAAUCAGGAUGCAGUGGAGAAUUUGCAAAUGCGCCCGGCGGCCAACUCCUUGCUCUUCC ....(((---(((((((.(((.((.(....).))(((((((..(((((.((((((........)))))).))).))...))).))))...)))))))))))))................. ( -37.30) >DroMoj_CAF1 299 120 + 1 AUCAAAUGAUGCUGGGCGGCAAGGAGUUGGCGCCACGUUUCAAGCGCAACUUGAUAUCGACCAAUCAGGAUUCGGUUGAGAAUCUGCAAAUGCGUCCAGCAGCUAAUUCGUUGCUGUUCC .............(((((((((.((((((((((....((((((.((...((((((........))))))...)).))))))....))...(((.....))))))))))).))))))))). ( -43.40) >consensus AUCAGCU___GCUGGGCAGCAAGGAGUUGGCGCCGCGUUUCAAGCGCAAUUUGAUCACCACGAAUCAGGAUGCGGUGGAGAAUCUGCAAAUGCGCCCGGCAGCCAACUCCUUGCUGUUCC .(((((....)))))..(((((.((((((((((((.(......(((((.((((((........)))))).....(..(.....)..)...)))))))))).)))))))).)))))..... (-27.81 = -29.45 + 1.64)

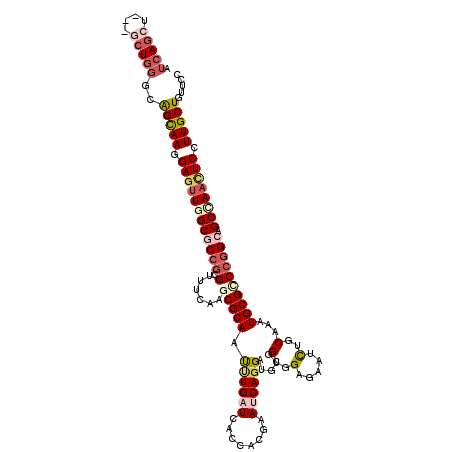
| Location | 8,437,130 – 8,437,247 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -43.75 |
| Consensus MFE | -30.49 |
| Energy contribution | -30.52 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8437130 117 - 20766785 GGAAGAGCAAGGAGUUGGCUGCCGGGCGCAUUUGCAAGUUCUCCACCGCAUCCUGGUUCGUGGUGAUCAAGUUGCGUUUGAACCUCGGUGCCAAUUCCUUACUACCCAGC---AGCUGAU ((...((.(((((((((((.((((((..((..(((((.((...((((((..........))))))...)).)))))..))..)).))))))))))))))).))..))...---....... ( -47.60) >DroVir_CAF1 6947 120 - 1 GGAACAGCAACGAGUUGGCCGCCGGGCGCAUUUGCAGAUUCUCAACCGAAUCCUGAUUGGUGGUGAUCAGAUUGCGUUUAAAGCGCGGCGCCAACUCCUUGCCGCCCAGCAUCAGCUGAU ((....((((.((((((((.((((.((.....(((((((((......)))))((((((......))))))..))))......)).)))))))))))).))))..))((((....)))).. ( -51.90) >DroPse_CAF1 25583 117 - 1 GGAAAAGCAAGGAGUUGGCCGCCGGGCGCAUUUGCAAGUUUUCCACCGCAUCCUGGUUCGUGGUGAUCAAAUUGCGCUUGAAACGCGGCGCUAGCUCCUUGCUGCCCAGC---AGCUGAU ((...((((((((((((((.((((.(..((..(((((......((((((..........))))))......)))))..))...).))))))))))))))))))..))...---....... ( -52.00) >DroGri_CAF1 313 117 - 1 GGAACAGCAACGAGUUGGCUGCCGGACGCAUCUGCAGAUUCUCAACCGAAUCCUGAUUCGCUGUGAUCAUAUUGCGCUUGAAACGCGGCGCCAACUCUUUGCCA---AGGAGCAUCUGAU ......((((.((((((((.(((((...((..(((((((..(((..(((((....)))))...)))..)).)))))..))...).)))))))))))).))))..---............. ( -38.30) >DroWil_CAF1 267 117 - 1 GGAAGAGCAAGGAGUUGGCCGCCGGGCGCAUUUGCAAAUUCUCCACUGCAUCCUGAUUUGUGGUGAUCAAAUUGCGCUUGAAUCGCGGUGCUAACUCCUUACUGCCCAGC---AGCUGAU .....((((((((((((((.((((((..((..(((((......(((..((........))..)))......)))))..))..)).)))))))))))))))..(((...))---))))... ( -42.20) >DroMoj_CAF1 299 120 - 1 GGAACAGCAACGAAUUAGCUGCUGGACGCAUUUGCAGAUUCUCAACCGAAUCCUGAUUGGUCGAUAUCAAGUUGCGCUUGAAACGUGGCGCCAACUCCUUGCCGCCCAGCAUCAUUUGAU ....(((((.(......).)))))((.((.....(((((((......)))).)))..(((.((.((...(((((((((........)))).)))))...)).)).))))).))....... ( -30.50) >consensus GGAACAGCAACGAGUUGGCCGCCGGGCGCAUUUGCAAAUUCUCAACCGAAUCCUGAUUCGUGGUGAUCAAAUUGCGCUUGAAACGCGGCGCCAACUCCUUGCCGCCCAGC___AGCUGAU ......((((.((((((((.(((((...((..(((((..((.((((.............)))).)).....)))))..))...).)))))))))))).))))....((((....)))).. (-30.49 = -30.52 + 0.03)
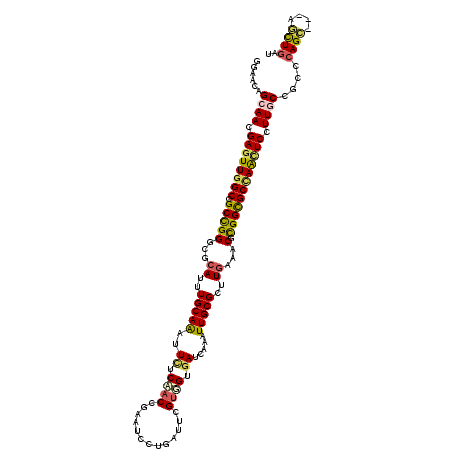

| Location | 8,437,207 – 8,437,317 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -28.74 |
| Energy contribution | -28.10 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8437207 110 - 20766785 CUGCCGC---UGCUAUUGGGCGGUGUUGACAUGGGCAACGUAGCCGGAAACUUUUGGUUCUGGGAGGCCGCUCGGAAGAGCAAGGAGUUGGCUGCCGGGCGCAUUUGCAAGUU .((((((---((((....))))))).....(((.((...(((((((...((((((((((......))))((((....)))).)))))))))))))...)).)))..))).... ( -42.60) >DroVir_CAF1 7027 110 - 1 CUGGAGU---UGCCGCUGGGCGGCGUCGAGAUGGGCAGCGUGGCCGGGAAUUUUUGAUUCUGUGAGGCGGCACGGAACAGCAACGAGUUGGCCGCCGGGCGCAUUUGCAGAUU (((.((.---(((.(((.((((((((((((((.(((......))).)....)))))))((((((......)))))).((((.....)))))))))).)))))).)).)))... ( -45.20) >DroWil_CAF1 344 113 - 1 UUGCUGCCGCCACCAUUUGGUGGUGUCGAUAUAGGCAAGUUUGCUGGGAAUUUUUGAUUUUGCGAUGCCGCUCGGAAGAGCAAGGAGUUGGCCGCCGGGCGCAUUUGCAAAUU .(((.(((((((((....))))))((((((...((((...((((...(((((...))))).))))))))((((....)))).....)))))).....)))))).......... ( -40.90) >DroMoj_CAF1 379 110 - 1 CUGCUGU---UGCCGCUGGGCGGCGUCGAAAUGGGCAGCGUGGCCGGGAAUUUCUGAUUCUGCGAGGCGGCACGGAACAGCAACGAAUUAGCUGCUGGACGCAUUUGCAGAUU ((((...---(((((((..((((.((((((((.(((......)))....))))).))).))))..)))))))((...(((((.(......).)))))..)).....))))... ( -43.40) >DroAna_CAF1 331 110 - 1 UUGCUGU---UGCUGUUCGGCGGCGUGGAUAUGGGCAGCGUGGCUGGGAACUUUUGGUUCUGGGAGGCCGCUCGGAAGAGCAAGGAGUUGGCUGCCGGGCGCAUCUGCAAGUU ((((...---(((.((((((((((.........(((.......((.(((((.....))))).))..)))((((....)))).........)))))))))))))...))))... ( -48.60) >DroPer_CAF1 23895 105 - 1 -----GC---UGCUGUUGGGUGGCGUCGACAUGGGCAACGUGGCUGGGAACUUCUGGUUUUGGUUGGCCGCUCGGAAAAGCAAGGAGUUGGCCGCCGGGCGCAUUUGCAAGUU -----((---(((.(((.((((((..((((..((.((((..(((..((....))..)))...)))).))(((......))).....)))))))))).))))))...))..... ( -38.50) >consensus CUGCUGC___UGCCGUUGGGCGGCGUCGACAUGGGCAACGUGGCCGGGAACUUUUGAUUCUGGGAGGCCGCUCGGAAGAGCAAGGAGUUGGCCGCCGGGCGCAUUUGCAAAUU .....((...(((.(((.((((((.........(((......)))...(((((((..(((((((......))))))).....))))))).)))))).))))))...))..... (-28.74 = -28.10 + -0.64)
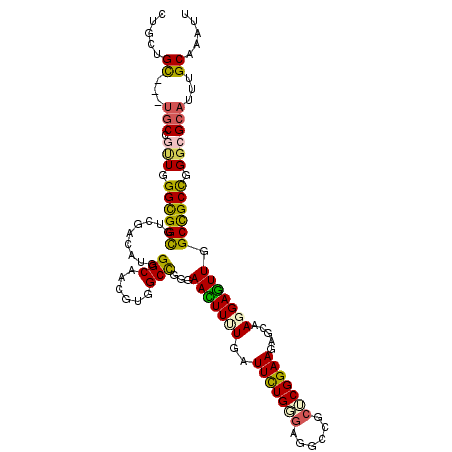
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:54 2006