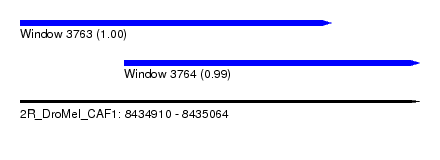
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,434,910 – 8,435,064 |
| Length | 154 |
| Max. P | 0.999953 |

| Location | 8,434,910 – 8,435,030 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -37.14 |
| Energy contribution | -37.38 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.82 |
| SVM RNA-class probability | 0.999953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
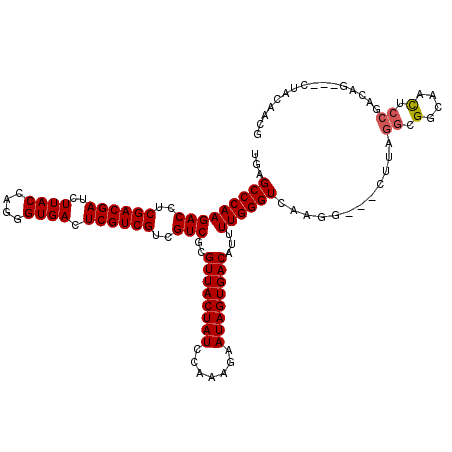
>2R_DroMel_CAF1 8434910 120 + 20766785 AGUAAGAAUUCCGUCAAAAUUCGCCAACACGACAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCGCGUUACUAUCCAAAGAAUAGUGACAUUUUGGGU ....................(((((..(((....))).)))))(((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...)))))) ( -40.60) >DroSec_CAF1 45905 120 + 1 AGCAAGAAUUCCGUCAAAAUUCGCCAACACGACAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCGCGUUACUAUCCAAAGAAUAGUGACAUUUUGGGU ....................(((((..(((....))).)))))(((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...)))))) ( -40.60) >DroSim_CAF1 45688 120 + 1 AGCAAGAAUUCCGUCAAAAUUCGCCAACACGACAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCGCGUUACUAUCCAAAGAAUAGUGACAUUUUGGGU ....................(((((..(((....))).)))))(((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...)))))) ( -40.60) >DroWil_CAF1 68439 120 + 1 AGCAAGAACUCCGCCAAAAUUCGUCAACACGAUAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCGCGUUACUAUAACAAAAAUAGUGACUUAUUGGGU ...........((((..(..((((....))))...)..)))).(((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...)))))) ( -39.20) >DroAna_CAF1 47238 120 + 1 AAUAAGAAUUCCGCCAAAAUUCGCCAACACGAUAGCGAAGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGAUUCGUCGUCGUCGCGUUACUAUCAAAAAAAUAGUGACAUUUUGGGU ..............((...(((((..........))))).)).(((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...)))))) ( -37.50) >consensus AGCAAGAAUUCCGUCAAAAUUCGCCAACACGACAGUGAGGUGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCGCGUUACUAUCCAAAGAAUAGUGACAUUUUGGGU ....................(((((..(((....))).)))))(((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...)))))) (-37.14 = -37.38 + 0.24)


| Location | 8,434,950 – 8,435,064 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.21 |
| Mean single sequence MFE | -43.24 |
| Consensus MFE | -33.54 |
| Energy contribution | -33.78 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8434950 114 + 20766785 UGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCGCGUUACUAUCCAAAGAAUAGUGACAUUUUGGGUCAAGG---CUUAGGCGGCAACUCCGACAG---CUACAACG (((.((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...)))))))).((---((..((.(....).))...))---))...... ( -41.80) >DroSec_CAF1 45945 114 + 1 UGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCGCGUUACUAUCCAAAGAAUAGUGACAUUUUGGGUCAAGG---CUUAGGCGGCAACUCCGAAAG---CUACAACG (((.((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...)))))))).((---(((.((.(....).))..)))---))...... ( -42.80) >DroSim_CAF1 45728 114 + 1 UGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCGCGUUACUAUCCAAAGAAUAGUGACAUUUUGGGUCAAGG---CUUAGGCGGCAACUCCGAAAG---CUACAACG (((.((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...)))))))).((---(((.((.(....).))..)))---))...... ( -42.80) >DroWil_CAF1 68479 110 + 1 UGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCGCGUUACUAUAACAAAAAUAGUGACUUAUUGGGUACAGGAGGUGCAGGCGGCAGCA---GCAG---CU----CG ((.(((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...)))))).)).(((.(((..((....)).---))).---))----). ( -44.60) >DroAna_CAF1 47278 120 + 1 UGAGCCCAAGACCUCGACGAUCUUACCAGGGUGAUUCGUCGUCGUCGCGUUACUAUCAAAAAAAUAGUGACAUUUUGGGUCAAGGCGGCAUCGGCGGUAAUACCGACAACGUCUACAACG (((.((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...))))))))(((((...((((........))))...)))))...... ( -44.20) >consensus UGAGCCCAAGACCUCGACGAUCUUACCAGGGUGACUCGUCGUCGUCGCGUUACUAUCCAAAGAAUAGUGACAUUUUGGGUCAAGG___CUUAGGCGGCAACUCCGACAG___CUACAACG ...(((((((((..((((((..((((....)))).))))))..)))..((((((((.......))))))))...))))))............((.(....).))................ (-33.54 = -33.78 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:51 2006