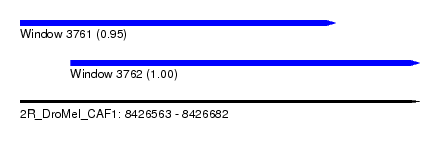
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,426,563 – 8,426,682 |
| Length | 119 |
| Max. P | 0.996385 |

| Location | 8,426,563 – 8,426,657 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -18.70 |
| Consensus MFE | -16.84 |
| Energy contribution | -17.36 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8426563 94 + 20766785 AUGAGAAUUCUUAUAAUCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUCAAAAACCACUAGCAUUCCAUUUCGAUGUGACAGAUUCCAAACCUAA (((((....)))))............((((.((((((((((((((((...........))))))))....))).))))).)))).......... ( -19.10) >DroSec_CAF1 37791 94 + 1 AUGAGAAUUCUUAUAAUCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUCAAAAACCACUAGCAUUCCAUUUCGAUGUGACAGAUUCCAAGCCUAA (((((....)))))............((((.((((((((((((((((...........))))))))....))).))))).)))).......... ( -19.10) >DroSim_CAF1 37246 94 + 1 AUAAGAAUUCUUAUAAUCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUCAAAAACCACUAGCAUUCCAUUUCGAUGUGACAGAUUCCAAGCCUAA (((((....)))))............((((.((((((((((((((((...........))))))))....))).))))).)))).......... ( -19.00) >DroEre_CAF1 40756 94 + 1 AUCAGAAUUCUUAUAAUCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUCAGAAAGCACUAGCAUUCCAUUUCGAUGUGACAGAUUCCAAACCUAA ....(((((.......((....)).......((((((((((((((((.((......))))))))))....))).))))).)))))......... ( -19.30) >DroYak_CAF1 39079 94 + 1 AUCAGAAUUCUUAUAACCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUUAAAAAGCACUAGCAUUCCAUUUCAAUGUGACAGAUUCCAAACCUAA ....(((((.(((((((((..........))))......((((((((...........)))))))).......)))))..)))))......... ( -17.00) >consensus AUCAGAAUUCUUAUAAUCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUCAAAAACCACUAGCAUUCCAUUUCGAUGUGACAGAUUCCAAACCUAA (((((....)))))............((((.((((((((((((((((...........))))))))....))).))))).)))).......... (-16.84 = -17.36 + 0.52)


| Location | 8,426,578 – 8,426,682 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 91.34 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -20.94 |
| Energy contribution | -21.34 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8426578 104 + 20766785 AUCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUCAAAAACCACUAGCAUUCCAUUUCGAUGUGACAGAUUCCAAACCUAAUGAGAAAAUUUAAUUGGUUGGUUCA .(((((((...((((.((((((((((((((((...........))))))))....))).))))).))))......)))))))...................... ( -22.90) >DroSec_CAF1 37806 100 + 1 AUCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUCAAAAACCACUAGCAUUCCAUUUCGAUGUGACAGAUUCCAAGCCUAAUGAGAAAAUUUAAUUG----GUCCA .(((((((...((((.((((((((((((((((...........))))))))....))).))))).))))......))))))).............----..... ( -22.90) >DroSim_CAF1 37261 100 + 1 AUCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUCAAAAACCACUAGCAUUCCAUUUCGAUGUGACAGAUUCCAAGCCUAAUGAGAAAAUUUAAUUG----GUCCA .(((((((...((((.((((((((((((((((...........))))))))....))).))))).))))......))))))).............----..... ( -22.90) >DroEre_CAF1 40771 100 + 1 AUCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUCAGAAAGCACUAGCAUUCCAUUUCGAUGUGACAGAUUCCAAACCUAAUGACAACAUUUAUUUU----CUACG .(((((((...((((.((((((((((((((((.((......))))))))))....))).))))).))))......))))))).............----..... ( -24.50) >DroYak_CAF1 39094 100 + 1 ACCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUUAAAAAGCACUAGCAUUCCAUUUCAAUGUGACAGAUUCCAAACCUAAUGAGAACAUUUAUUUC----UUUCG ..((((((...((((.(((((...((((((((...........))))))))........))))).))))......))))))..............----..... ( -20.10) >consensus AUCAUUAGAAAAAUUGGUCAUUCGGGAGUGCUCAAAAACCACUAGCAUUCCAUUUCGAUGUGACAGAUUCCAAACCUAAUGAGAAAAUUUAAUUG____GUCCA .(((((((...((((.((((((((((((((((...........))))))))....))).))))).))))......)))))))...................... (-20.94 = -21.34 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:49 2006