
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,424,198 – 8,424,292 |
| Length | 94 |
| Max. P | 0.951427 |

| Location | 8,424,198 – 8,424,292 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.98 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.22 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
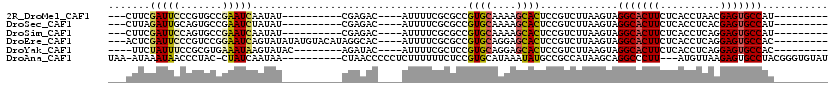
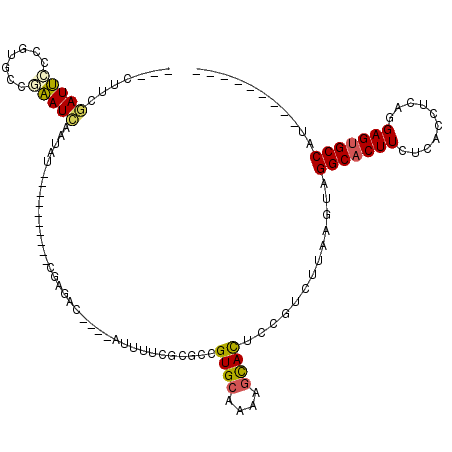
>2R_DroMel_CAF1 8424198 94 - 20766785 ---CUUCGAUUCCCGUGCCGAAUCAAUAU----------CGAGAC----AUUUUCGCGCCGUGCAAAAGCACUCCGUCUUAAGUAGGCACUUCUCACCUAACGAGUGCCAU--------- ---....(((((.......))))).....----------......----((((..(((..((((....))))..)))...)))).(((((((..........)))))))..--------- ( -21.40) >DroSec_CAF1 35471 94 - 1 ---CUUAGAUUGCAGUGCCGAAUCUAUAU----------CGAGAC----AUUUUCGCGCCGUGCAAAAGCACUCCGUCUUAAGUAGGCACUUCUCACCUCACGAGUGCCAU--------- ---(((((((....((((.((((((....----------..))).----...))))))).((((....))))...))).))))..(((((((..........)))))))..--------- ( -22.60) >DroSim_CAF1 35803 94 - 1 ---CUUCGAUUCCAGUGCCGAAUCAAUAU----------CGAGAC----AUUUUCGCGCCGUGCAAAAGCACUCCGUCUUAAGUAGGCACUUCUCACCUCAGGAGUGCCAU--------- ---....(((((.......))))).....----------......----((((..(((..((((....))))..)))...)))).(((((((((......)))))))))..--------- ( -26.10) >DroEre_CAF1 38217 104 - 1 ---ACUCGAUUCCCGUCCGGAAUCAGUAUAUAUGUACAUAGGCAC----AUUUUCGCGCCGUGCAGGAGCACUCCGUCUUAAGUAGGCACUUCUCACCUCAGGAGUGCCAC--------- ---(((.((((((.....))))))))).....(((((...(((.(----......).))))))))(((....)))..........(((((((((......)))))))))..--------- ( -34.40) >DroYak_CAF1 36560 95 - 1 ----UUCUAUUUCCGCGUGAAAUAAGUAUAC--------AGAUAC----AUUUUCGCUCCGUGCAGGAGCACUCCGUCUUAAGUAGGCACUUCUCACCUCAGGAGUGCCAC--------- ----...(((((..(((.((((...((((..--------..))))----..))))(((((.....)))))....)))...)))))(((((((((......)))))))))..--------- ( -29.70) >DroAna_CAF1 35519 105 - 1 UAA-AUAAAUAACCCUAC-CUAUCAAUAA----------CUAACCCCCUCUUUUUUCUCCGUGCAUAAAUAUGCCGCCAUAAGCAGGCCCUU---AUGUUAAGAGUGCCUACGGGUGUAU ...-.......((((...-..........----------....................((.((((....))))))......(.((((.(((---.......))).)))).))))).... ( -17.40) >consensus ___CUUCGAUUCCCGUGCCGAAUCAAUAU__________CGAGAC____AUUUUCGCGCCGUGCAAAAGCACUCCGUCUUAAGUAGGCACUUCUCACCUCAGGAGUGCCAU_________ .......(((((.......)))))....................................((((....)))).............(((((((..........)))))))........... (-15.32 = -15.22 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:47 2006