
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,410,989 – 8,411,080 |
| Length | 91 |
| Max. P | 0.966563 |

| Location | 8,410,989 – 8,411,080 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.63 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -13.17 |
| Energy contribution | -14.53 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
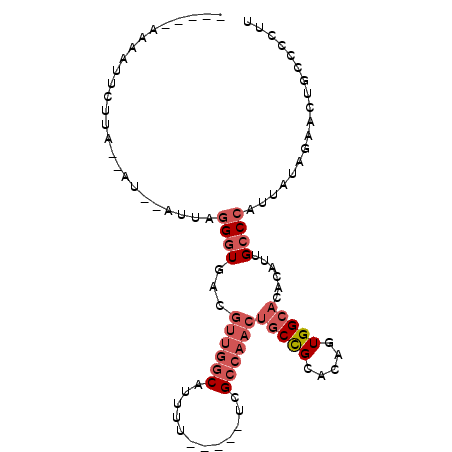
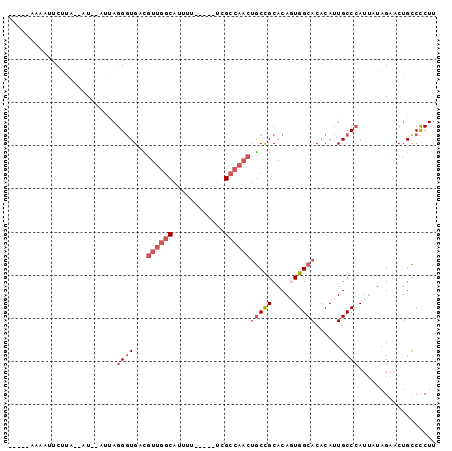
>2R_DroMel_CAF1 8410989 91 + 20766785 UCGGGAAAAUUCUUA--UU--AUUAGGGUGACGUUGGCAUUUUU----UCGCCAACUGCCGCACAGUGGCACACAUUGCCCAUUUUAGAACUGCCCCUU ..(((..(.((((..--..--....(((..(.((((((......----..))))))((((((...))))))....)..))).....)))).)..))).. ( -29.32) >DroPse_CAF1 21853 84 + 1 -----UAAAUUCUUCCCGU--AACAGGGUGCUCUUGGCAUUU--GCUUCUGC-----ACUGCACAGUGGCACACAUUGCCGAUUAUAGA-CGGUCCCUU -----..............--...((((((((.((((((..(--((....))-----).))).))).))))).....((((........-)))).))). ( -21.50) >DroSec_CAF1 22514 91 + 1 UCAGGAAAAUUCUUA--AU--AUCAGGGUGACGUUGACAUUUUU----UCGCCAACUGCCGCACACUGGCACACAUUGCCCAUUUUAGAACUGCCCCUU ...((....((((..--..--....(((..(.((((.(......----..).))))(((((.....)))))....)..))).....))))...)).... ( -16.42) >DroSim_CAF1 22146 91 + 1 UCAGCAAAAUUCUUA--UU--AUUAGGGUGACGUUGGCAUUUUU----UCGCCAACUGCCGCACACUGGCACACAUUGCCCAUUUUAGAACUGCCCCUU ...(((...((((..--..--....(((..(.((((((......----..))))))(((((.....)))))....)..))).....)))).)))..... ( -23.92) >DroEre_CAF1 24389 80 + 1 ----------UCUUA--AU--AUUAGGGUGUCGUUGGCAUUUU-----UCGCCAACUGCCGCACAGUGGCACACAUUGCCCAUUAUAGAACUGCCCCUU ----------.....--..--....((((...((((((.....-----..))))))((((((...))))))......)))).................. ( -22.90) >DroYak_CAF1 22420 81 + 1 -----------CCAA--AUGCAGUAGGCUGCUGUUGGCAUUUU-----UUGCCAACUGCCGCACAGUGGCACACAUUGCCCAUAAUAGAACUGCCUCUU -----------....--..(((((..((.((.(((((((....-----.))))))).)).))...(.((((.....)))))........)))))..... ( -28.50) >consensus _____AAAAUUCUUA__AU__AUUAGGGUGACGUUGGCAUUUU_____UCGCCAACUGCCGCACAGUGGCACACAUUGCCCAUUAUAGAACUGCCCCUU .........................((((...((((((............))))))(((((.....)))))......)))).................. (-13.17 = -14.53 + 1.36)

| Location | 8,410,989 – 8,411,080 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 76.63 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.46 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
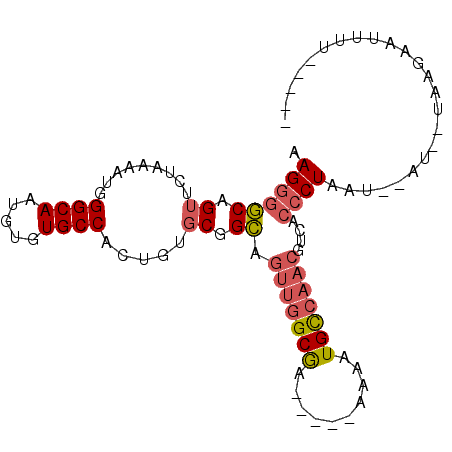
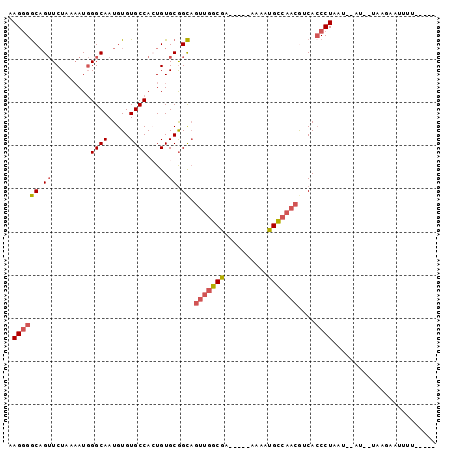
>2R_DroMel_CAF1 8410989 91 - 20766785 AAGGGGCAGUUCUAAAAUGGGCAAUGUGUGCCACUGUGCGGCAGUUGGCGA----AAAAAUGCCAACGUCACCCUAAU--AA--UAAGAAUUUUCCCGA ..((((.((((((......((((.....)))).......(((.(((((((.----.....))))))))))........--..--..)))))).)))).. ( -29.10) >DroPse_CAF1 21853 84 - 1 AAGGGACCG-UCUAUAAUCGGCAAUGUGUGCCACUGUGCAGU-----GCAGAAGC--AAAUGCCAAGAGCACCCUGUU--ACGGGAAGAAUUUA----- .........-(((......((((.....(((..((((.....-----))))..))--)..)))).......(((....--..))).))).....----- ( -17.90) >DroSec_CAF1 22514 91 - 1 AAGGGGCAGUUCUAAAAUGGGCAAUGUGUGCCAGUGUGCGGCAGUUGGCGA----AAAAAUGUCAACGUCACCCUGAU--AU--UAAGAAUUUUCCUGA ..((((.((((((......((((.....)))).......(((.(((((((.----.....))))))))))........--..--..)))))).)))).. ( -23.80) >DroSim_CAF1 22146 91 - 1 AAGGGGCAGUUCUAAAAUGGGCAAUGUGUGCCAGUGUGCGGCAGUUGGCGA----AAAAAUGCCAACGUCACCCUAAU--AA--UAAGAAUUUUGCUGA .(((((((...(......)((((.....))))....)))(((.(((((((.----.....)))))))))).))))...--..--............... ( -25.00) >DroEre_CAF1 24389 80 - 1 AAGGGGCAGUUCUAUAAUGGGCAAUGUGUGCCACUGUGCGGCAGUUGGCGA-----AAAAUGCCAACGACACCCUAAU--AU--UAAGA---------- .((((...((..((((...((((.....))))..))))..)).(((((((.-----....)))))))....))))...--..--.....---------- ( -24.50) >DroYak_CAF1 22420 81 - 1 AAGAGGCAGUUCUAUUAUGGGCAAUGUGUGCCACUGUGCGGCAGUUGGCAA-----AAAAUGCCAACAGCAGCCUACUGCAU--UUGG----------- .....(((((........((((......((((.......))))(((((((.-----....)))))))....)))))))))..--....----------- ( -27.40) >consensus AAGGGGCAGUUCUAAAAUGGGCAAUGUGUGCCACUGUGCGGCAGUUGGCGA_____AAAAUGCCAACGUCACCCUAAU__AU__UAAGAAUUUU_____ .((((((.((.........((((.....)))).....)).)).(((((((..........)))))))....))))........................ (-16.93 = -17.46 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:44 2006