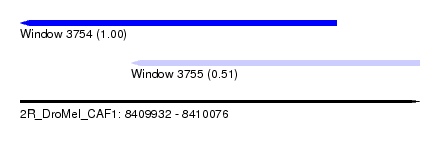
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,409,932 – 8,410,076 |
| Length | 144 |
| Max. P | 0.999450 |

| Location | 8,409,932 – 8,410,046 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -26.93 |
| Energy contribution | -26.68 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
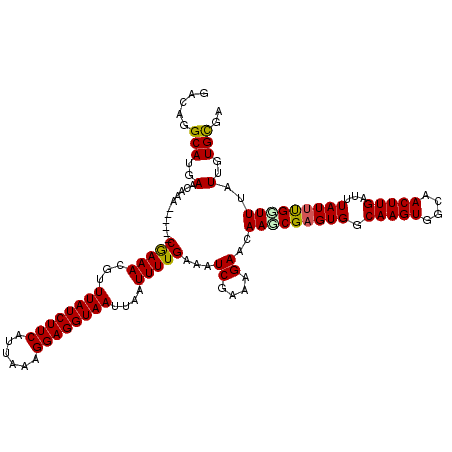
>2R_DroMel_CAF1 8409932 114 - 20766785 GACAGGCAUGAACAAA------CGAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAAGCGAGUGGCAAGUGGCAACUUGAUUUAUUUGUUUUAUUGUGCGC .....(((..(.....------(((((...((((((((......))))))))....)))))...((....))..(((((((((.(((((....)))))...)))))))))..)..))).. ( -27.10) >DroVir_CAF1 26795 120 - 1 GACAGGCAUGAACAAACAUGAACAAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAACCGAGUGGCAAGUGGCAACUUGAUUUAUUUGGUUUAUUGUGCGA .....(((..(...........(((((...((((((((......))))))))....)))))...((....))..(((((((((.(((((....)))))...)))))))))..)..))).. ( -29.80) >DroGri_CAF1 28828 114 - 1 GACAGGCAUGAACAAA------CGAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAACCGAGUGGCAAGUGGCAACUUGAUUUAUUCGGUUUAUUGUGCGA .....(((..(.....------(((((...((((((((......))))))))....)))))...((....))..(((((((((.(((((....)))))...)))))))))..)..))).. ( -31.60) >DroWil_CAF1 26871 114 - 1 GACAGGCAUGAACAAA------CGAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAAGCGAGUGGCAAGUGGCAACUUGAUUUAUUUGGUUUAUUGUGUGC ..((.((((((((...------(((((...((((((((......))))))))....)))))...((....)).....((((((.(((((....)))))...))))))))))).))).)). ( -26.80) >DroMoj_CAF1 30505 114 - 1 GACAAGCAUGAACAAA------CAAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAAGCGAGUGGCAAGUGGCAACUUGAUUUAUUUGGUUUAUUGUGCGA .....(((..(.....------(((((...((((((((......))))))))....)))))((((((((.(((....(((((.((.....)).))))).))).)))))))).)..))).. ( -26.60) >DroPer_CAF1 20695 114 - 1 GACAGGCAUGAACAAA------CGAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAAGCGAGUGGCAAGUGGCAACUUGAUUUAUAUGCUUUAUUGUGCGC .....(((..(.....------(((((...((((((((......))))))))....)))))...((....))..(((((.(((.(((((....)))))...))).)))))..)..))).. ( -25.90) >consensus GACAGGCAUGAACAAA______CGAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAAGCGAGUGGCAAGUGGCAACUUGAUUUAUUUGGUUUAUUGUGCGA .....(((..(...........(((((...((((((((......))))))))....)))))...((....))..(((((((((.(((((....)))))...)))))))))..)..))).. (-26.93 = -26.68 + -0.25)

| Location | 8,409,972 – 8,410,076 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.21 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -12.77 |
| Energy contribution | -12.88 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
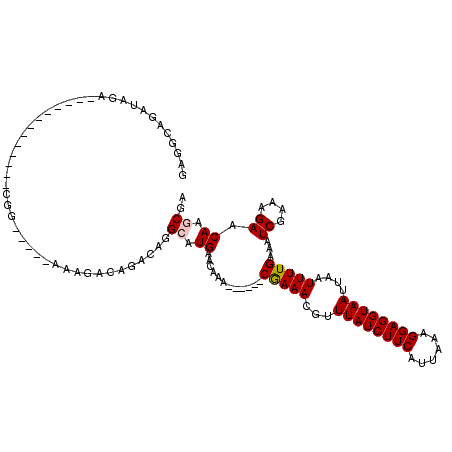
>2R_DroMel_CAF1 8409972 104 - 20766785 GCAGCAGAUAGA----------CGACGGACAGGCAAAACAGACAGGCAUGAACAAA------CGAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAAGCGA ...((.......----------....(..((.((...........)).))..)...------(((((...((((((((......))))))))....)))))...((....))....)).. ( -15.30) >DroVir_CAF1 26835 94 - 1 GGGGCGAG-------------------------CA-GAGCGACAGGCAUGAACAAACAUGAACAAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAACCGA (..((...-------------------------..-..))..).((((((......))))..(((((...((((((((......))))))))....)))))...((....))....)).. ( -17.40) >DroGri_CAF1 28868 94 - 1 AAGG--GGUUGG-------------GGG-----AGAGUGCGACAGGCAUGAACAAA------CGAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAACCGA ....--(((((.-------------...-----...((((.....)))).......------(((((...((((((((......))))))))....)))))...((....)).))))).. ( -21.80) >DroWil_CAF1 26911 114 - 1 GACACAGAGAGACUAAAAUGAAUGCAGGAGAAGAUAGACAGACAGGCAUGAACAAA------CGAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAAGCGA ......................(((....((((((((((.....(...........------).....))))))))))..........................((....))....))). ( -15.90) >DroMoj_CAF1 30545 95 - 1 GCGGCUGGUUGA-------------GGG-----AA-CGGGGACAAGCAUGAACAAA------CAAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAAGCGA ((.(((.(((..-------------(..-----..-)...))).))).((......------(((((...((((((((......))))))))....)))))...((....)).)).)).. ( -19.80) >DroAna_CAF1 21267 101 - 1 GGAGCUGAUAGA-------------CGGACAGGCAAAGCAGACAGGCAUGAACAAA------CGAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAAGCGA ...(((......-------------(.....)....)))......((.((......------(((((...((((((((......))))))))....)))))...((....)).)).)).. ( -16.50) >consensus GAGGCAGAUAGA_____________CGG_____AAAGACAGACAGGCAUGAACAAA______CGAAACGUUUAUCUUCAUUAAAGGAGGUAAUUAAUUUUGAAAUCGAAAGAACAAGCGA .............................................((.((............(((((...((((((((......))))))))....)))))...((....)).)).)).. (-12.77 = -12.88 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:43 2006