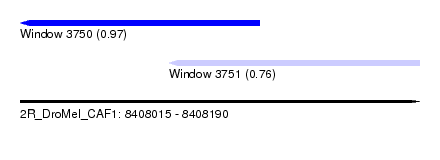
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,408,015 – 8,408,190 |
| Length | 175 |
| Max. P | 0.974917 |

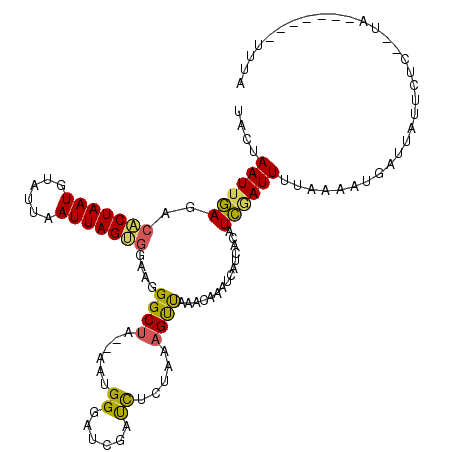
| Location | 8,408,015 – 8,408,120 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.50 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -8.40 |
| Energy contribution | -7.88 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8408015 105 - 20766785 UACUAAUUGAGACACUAAUGUAUUAAUUAGUGGAAGGGCUA--AAUGGAAACGAUCUCUAAAGUUAAACAAAUCAUCAUAUCUAUUUUAAAAUGAUUGUUCUC-----------UUUA ........(((((((((((......))))))).........--..((....)).))))(((((...(((((.((((..((.......))..)))))))))..)-----------)))) ( -18.80) >DroSec_CAF1 19510 104 - 1 UCCUAAUUGAGACACUAAUGUAUUAAUUAGUGGAAGGGCUA--AAUGGGUCCGAUCUCUAAAGUUAAAUAAAUCAACACAUCGAUUUUUAAAAGAUUAUUCU------------UCUC ........(((((((((((......)))))))((((((((.--....)))))(((((......(((((..((((........))))))))).))))).))).------------)))) ( -19.70) >DroSim_CAF1 19130 116 - 1 UACUAAUUGAGACACUAAUGUAUUAAUUAGUGGAAGGGCUA--AAUGGGAUCGAUCUCUAAAGUUAAAUAAAUCAACACAUCGAUUUUUAAAAGAUUAUUCUCAUUAUAAGCUCUUUC .(((((((((.(((....))).)))))))))(((((((((.--(((((((..(((((......(((((..((((........))))))))).)))))..)))))))...))))))))) ( -29.90) >DroEre_CAF1 21434 115 - 1 -ACUAAUUAAGACCCUAAUGUAUUAAUUAGGAGAACGGCUA--AAUGGAAGCGACCUCUAAAGUUCAAGUAAUCAUCACAUUAAUUUAAUGAUUACUAUUUUGCUUAGCCAUUUCUUA -.((((((((.((......)).))))))))(((((.(((((--(..((((..((.((....)).)).(((((((((............))))))))).))))..))))))..))))). ( -26.30) >DroYak_CAF1 19360 118 - 1 UACUAAUUGAAACACUAAUGUUUUAAUUAGGGGAACGGCGGAGAACGGGAUCGAUUUAUAAAGCCAAACGAAUCAUCACAUCGAUUUAAUAAUUAUUUUUUUCCCUAGAAGCCUUUUA ..((((((((((((....))))))))))))(((((.(((..(((.((....)).))).....)))....(((((........))))).............)))))............. ( -25.10) >consensus UACUAAUUGAGACACUAAUGUAUUAAUUAGUGGAAGGGCUA__AAUGGGAUCGAUCUCUAAAGUUAAACAAAUCAUCACAUCGAUUUUAAAAUGAUUAUUCUC__UA_______UUUA ....((((((..(((((((......)))))))....((((......((......)).....))))...............))))))................................ ( -8.40 = -7.88 + -0.52)

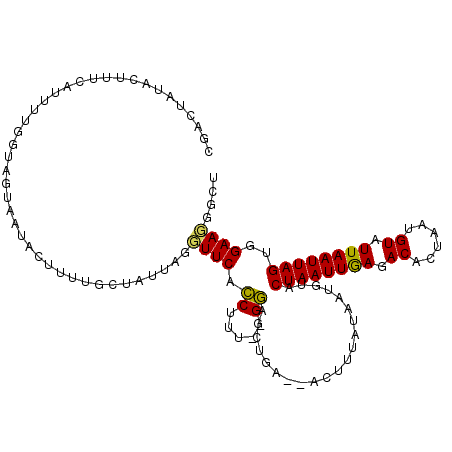
| Location | 8,408,080 – 8,408,190 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.64 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -9.84 |
| Energy contribution | -9.24 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8408080 110 - 20766785 CGACUAUACUUUAAUUUUGGUAGCAAUACUUUUGCUACUAUGUUCACCUUU--GGAGCUGA--ACUUUAUAAUGUACUAAUUGAGACACUAAUGUAUUAAUUAGUGGAAGGGCU ........((((.....(((((((((.....))))))))).(((((.((..--..)).)))--)).........((((((((((.(((....))).)))))))))))))).... ( -26.70) >DroSec_CAF1 19574 110 - 1 CGACUUUACUUUCAUUUUGGUAUUAAUACUUUUGCGAUUAGUUUCACCUUU--GGAGCUGA--ACUUUAUAAUGUCCUAAUUGAGACACUAAUGUAUUAAUUAGUGGAAGGGCU ........((((((((.....((((((((........(((((((((....)--))))))))--.........(((((.....).)))).....)))))))).)))))))).... ( -23.60) >DroSim_CAF1 19206 110 - 1 CGACUUUACUUUCAUUUUGGUAGUAAUACUUUUCAGAUUAGUUUCACCUUU--GGAGCUGA--ACUUUAUAAUGUACUAAUUGAGACACUAAUGUAUUAAUUAGUGGAAGGGCU ........(((((.(((..((((((.........((.(((((((((....)--))))))))--.))........)))).))..)))(((((((......))))))))))))... ( -26.13) >DroEre_CAF1 21510 107 - 1 AGCCUACACUUUCAUUUUGGUAGU-AUACUUUAGCUAUUAGGUUCAGCUCUAAGCUGA-----AGUGUAUAAUG-ACUAAUUAAGACCCUAAUGUAUUAAUUAGGAGAACGGCU ((((....(((.....(((((.((-(((((((((((..((((......))))))))))-----)))))))....-)))))..)))..((((((......)))))).....)))) ( -31.00) >DroYak_CAF1 19438 101 - 1 AGUCUAUACUUUCA-----------AUACUUUAGCUUUUAAGUUCACCUUU--GGUGACGAAUACUUUAUAAUGUACUAAUUGAAACACUAAUGUUUUAAUUAGGGGAACGGCG ..............-----------........(((.....((((.(((((--(....))))..............((((((((((((....))))))))))))))))))))). ( -22.40) >consensus CGACUAUACUUUCAUUUUGGUAGUAAUACUUUUGCUAUUAGGUUCACCUUU__GGAGCUGA__ACUUUAUAAUGUACUAAUUGAGACACUAAUGUAUUAAUUAGUGGAAGGGCU .........................................((((.((.....)).....................((((((((.((......)).))))))))..)))).... ( -9.84 = -9.24 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:39 2006