
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,407,433 – 8,407,567 |
| Length | 134 |
| Max. P | 0.999875 |

| Location | 8,407,433 – 8,407,547 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -54.72 |
| Consensus MFE | -42.62 |
| Energy contribution | -44.50 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8407433 114 + 20766785 GGCAGCUCCCACUCGCCCUGCAGCCCCAUCCUGCCGCCAUCGGUGAGCGCAAAUGCCGCUG---CGGCAGUGGCAGCGGCGGCCCACAAUAGUGCAGCGGCGGCAGUGGCGGUCGCA .((.(((.(((((.(((((((.(((.......((((((...)))).))......(((((((---(.......))))))))))).(((....)))..)))).)))))))).))).)). ( -54.60) >DroVir_CAF1 24133 114 + 1 GGCAGCUCUCAUUCGCCCUGCAGCCCCAUCCUGCCGCCCUCGGUGAGCGCCAAUGCAGCCGCAGCGGCUGUUGCCGCCGCC---CAUAACUCAGCGGCCGCAGCCGUUGCGGCGGUG (((.((((.(....((...((((.......)))).)).....).)))))))...((.((((((((((((((.(((((....---.........))))).)))))))))))))).)). ( -58.12) >DroGri_CAF1 26126 117 + 1 GGCAGCUCUCACUCCCCCUGCAGCCCCAUAUUGCCGCCGUCGGUGAGCGCCAAUGCGGCAGCAGCAGCUGUUGCCGCCGCCGCACACAAUUCAGCUGCAGCAGCCGUUGCAGCGGUU (((.((.((((((..(...((.((........)).)).)..)))))).))....((((((((((...)))))))))).)))............((((((((....)))))))).... ( -50.10) >DroWil_CAF1 22586 117 + 1 GGUAGCUCUCAUUCACCCUGCAGUCCCAUUCUACCGCCUUCGGUGAGCGCCAAUGCUGCAGCAGCAGCUGUUGCCGCCGCUGCACAUAAUUCGGCGGCGGCAGCUGUUGCUGCUGUG (((.((((.............((.......))((((....)))))))))))......(((((((((((.(((((((((((((.........))))))))))))).))))))))))). ( -58.30) >DroMoj_CAF1 27451 114 + 1 GGCAGCUCCCAUUCCCCCUGCAGCCCCAUUUUGCCGCCCUCGGUGAGCGCCAAUGCAGCGGCAGCGGCCGUUGCUGCCGCC---CACAAUUCGGCGGCUGCAGCCGUUGCGGCCGUU (((.((((.....((....((.((........)).))....)).))))))).....((((((.(((((.(((((.((((((---........)))))).))))).))))).)))))) ( -52.50) >consensus GGCAGCUCUCAUUCCCCCUGCAGCCCCAUCCUGCCGCCCUCGGUGAGCGCCAAUGCAGCAGCAGCGGCUGUUGCCGCCGCCGC_CACAAUUCAGCGGCGGCAGCCGUUGCGGCCGUG (((.((((...........((((.......)))).(((...))))))))))...((.((((((((((((((.(((((((............))))))).)))))))))))))).)). (-42.62 = -44.50 + 1.88)

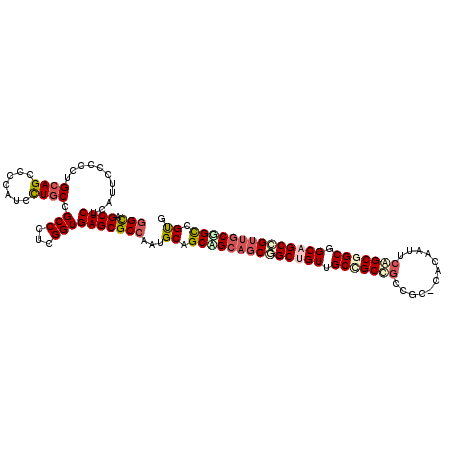

| Location | 8,407,433 – 8,407,547 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -56.86 |
| Consensus MFE | -35.20 |
| Energy contribution | -35.52 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8407433 114 - 20766785 UGCGACCGCCACUGCCGCCGCUGCACUAUUGUGGGCCGCCGCUGCCACUGCCG---CAGCGGCAUUUGCGCUCACCGAUGGCGGCAGGAUGGGGCUGCAGGGCGAGUGGGAGCUGCC ((((.((.((((((((((((((.(((....)))))).((((((((.......)---)))))))................))))))))..))))).)))).(((.(((....)))))) ( -55.00) >DroVir_CAF1 24133 114 - 1 CACCGCCGCAACGGCUGCGGCCGCUGAGUUAUG---GGCGGCGGCAACAGCCGCUGCGGCUGCAUUGGCGCUCACCGAGGGCGGCAGGAUGGGGCUGCAGGGCGAAUGAGAGCUGCC ...((((....((((((..(((((((.......---..)))))))..))))))((((((((.(((((.(((((.....))))).))..))).))))))))))))............. ( -61.40) >DroGri_CAF1 26126 117 - 1 AACCGCUGCAACGGCUGCUGCAGCUGAAUUGUGUGCGGCGGCGGCAACAGCUGCUGCUGCCGCAUUGGCGCUCACCGACGGCGGCAAUAUGGGGCUGCAGGGGGAGUGAGAGCUGCC ..((((.((....)).))(((((((.....(((.(((((((((((....))))))))))))))((((.((((.......)))).))))....)))))))))((.(((....))).)) ( -58.20) >DroWil_CAF1 22586 117 - 1 CACAGCAGCAACAGCUGCCGCCGCCGAAUUAUGUGCAGCGGCGGCAACAGCUGCUGCUGCAGCAUUGGCGCUCACCGAAGGCGGUAGAAUGGGACUGCAGGGUGAAUGAGAGCUACC ....((((((.((((((..(((((((............)))))))..)))))).))))))(((.((..((.(((((....(((((........)))))..))))).)).)))))... ( -53.30) >DroMoj_CAF1 27451 114 - 1 AACGGCCGCAACGGCUGCAGCCGCCGAAUUGUG---GGCGGCAGCAACGGCCGCUGCCGCUGCAUUGGCGCUCACCGAGGGCGGCAAAAUGGGGCUGCAGGGGGAAUGGGAGCUGCC ..(((((.((.(..((((((((.(((...((..---(.(((((((.......))))))))..))(((.(((((.....))))).)))..)))))))))))..)...)).).)))).. ( -56.40) >consensus AACCGCCGCAACGGCUGCCGCCGCCGAAUUGUG_GCGGCGGCGGCAACAGCCGCUGCUGCUGCAUUGGCGCUCACCGAGGGCGGCAGAAUGGGGCUGCAGGGCGAAUGAGAGCUGCC ....((.(((.((((((..((.((((............)))).))..)))))).))).)).....((((.((((((....(((((........))))).)).....)))).)))).. (-35.20 = -35.52 + 0.32)

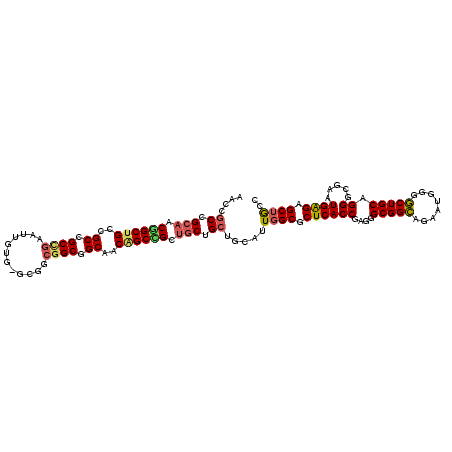
| Location | 8,407,473 – 8,407,567 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.17 |
| Mean single sequence MFE | -52.47 |
| Consensus MFE | -34.82 |
| Energy contribution | -36.70 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.66 |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8407473 94 + 20766785 CGGUGAGCGCAAAUGCCGCUG---CGGCAGUGGCAGCGGCGGCCCACAAUAGUGCAGCGGCGGCAGUGGCGGUCGCAGCGAAUCAGGCCAGCUCCUC---------- .((.((((......(((((((---(.......))))))))((((........(((.(((((.((....)).))))).))).....)))).)))))).---------- ( -46.42) >DroVir_CAF1 24173 94 + 1 CGGUGAGCGCCAAUGCAGCCGCAGCGGCUGUUGCCGCCGCC---CAUAACUCAGCGGCCGCAGCCGUUGCGGCGGUGGCCAAUCAGGCCUCCUCCUC---------- .(((....)))......((((((((((((((.(((((....---.........))))).))))))))))))))((.((((.....)))).)).....---------- ( -55.42) >DroGri_CAF1 26166 97 + 1 CGGUGAGCGCCAAUGCGGCAGCAGCAGCUGUUGCCGCCGCCGCACACAAUUCAGCUGCAGCAGCCGUUGCAGCGGUUGCGAAUCAAGCCUCCUCCUC---------- .((.(((.((....(((((.(((((....))))).)))))((((.((......((((((((....)))))))).))))))......)))))..))..---------- ( -43.30) >DroWil_CAF1 22626 107 + 1 CGGUGAGCGCCAAUGCUGCAGCAGCAGCUGUUGCCGCCGCUGCACAUAAUUCGGCGGCGGCAGCUGUUGCUGCUGUGGCAAAUCAGGCCAGUUCAUCCAGUUUGCAA .((((((((((..(((..((((((((((.(((((((((((((.........))))))))))))).))))))))))..))).....)))..))))))).......... ( -65.40) >DroMoj_CAF1 27491 94 + 1 CGGUGAGCGCCAAUGCAGCGGCAGCGGCCGUUGCUGCCGCC---CACAAUUCGGCGGCUGCAGCCGUUGCGGCCGUUGCGAAUCAAGCCUCCUCCUC---------- .((.(((.((...(((((((((.(((((.(((((.((((((---........)))))).))))).))))).)))))))))......)))))..))..---------- ( -51.80) >consensus CGGUGAGCGCCAAUGCAGCAGCAGCGGCUGUUGCCGCCGCCGC_CACAAUUCAGCGGCGGCAGCCGUUGCGGCCGUGGCGAAUCAGGCCUCCUCCUC__________ .(((((..((....((.((((((((((((((.(((((((............))))))).)))))))))))))).)).))...))..))).................. (-34.82 = -36.70 + 1.88)


| Location | 8,407,473 – 8,407,567 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.17 |
| Mean single sequence MFE | -46.50 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8407473 94 - 20766785 ----------GAGGAGCUGGCCUGAUUCGCUGCGACCGCCACUGCCGCCGCUGCACUAUUGUGGGCCGCCGCUGCCACUGCCG---CAGCGGCAUUUGCGCUCACCG ----------(..((((.(((((.....((.(((.(.((....)).).))).))((....)))))))((((((((.......)---)))))))......))))..). ( -41.90) >DroVir_CAF1 24173 94 - 1 ----------GAGGAGGAGGCCUGAUUGGCCACCGCCGCAACGGCUGCGGCCGCUGAGUUAUG---GGCGGCGGCAACAGCCGCUGCGGCUGCAUUGGCGCUCACCG ----------(((..((.((((.....)))).))((((((.((((((..(((((((.......---..)))))))..)))))).)))))).((....)).))).... ( -54.70) >DroGri_CAF1 26166 97 - 1 ----------GAGGAGGAGGCUUGAUUCGCAACCGCUGCAACGGCUGCUGCAGCUGAAUUGUGUGCGGCGGCGGCAACAGCUGCUGCUGCCGCAUUGGCGCUCACCG ----------..((..((((((.((((((.....((((((........))))))))))))..((((((((((((((.....)))))))))))))).))).))).)). ( -46.40) >DroWil_CAF1 22626 107 - 1 UUGCAAACUGGAUGAACUGGCCUGAUUUGCCACAGCAGCAACAGCUGCCGCCGCCGAAUUAUGUGCAGCGGCGGCAACAGCUGCUGCUGCAGCAUUGGCGCUCACCG .........((.(((..((((.......))))..(((((....))))).((.(((((....((((((((((((((....)))))))))))).)))))))))))))). ( -46.60) >DroMoj_CAF1 27491 94 - 1 ----------GAGGAGGAGGCUUGAUUCGCAACGGCCGCAACGGCUGCAGCCGCCGAAUUGUG---GGCGGCAGCAACGGCCGCUGCCGCUGCAUUGGCGCUCACCG ----------..((..(((.........((..((((((...))))))..))((((((..((..---(.(((((((.......))))))))..))))))))))).)). ( -42.90) >consensus __________GAGGAGGAGGCCUGAUUCGCAACCGCCGCAACGGCUGCCGCCGCCGAAUUGUG_GCGGCGGCGGCAACAGCCGCUGCUGCUGCAUUGGCGCUCACCG ............((....((((..(((((((......(((.((((((..((.((((............)))).))..)))))).)))))))).))..).)))..)). (-23.98 = -24.90 + 0.92)

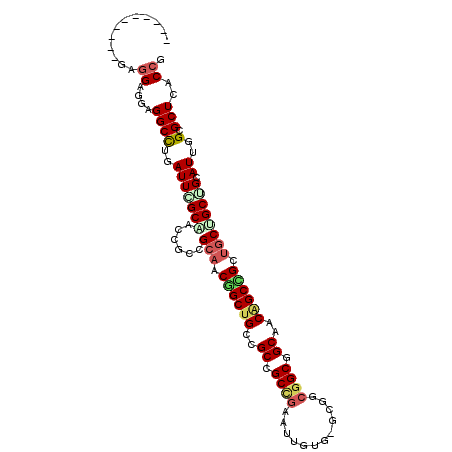
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:37 2006