
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,391,616 – 8,391,723 |
| Length | 107 |
| Max. P | 0.789147 |

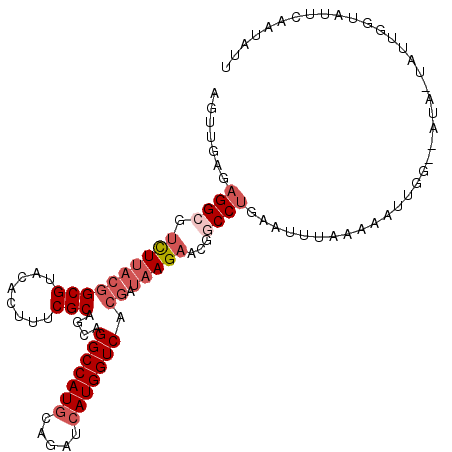
| Location | 8,391,616 – 8,391,723 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.33 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -18.47 |
| Energy contribution | -19.83 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
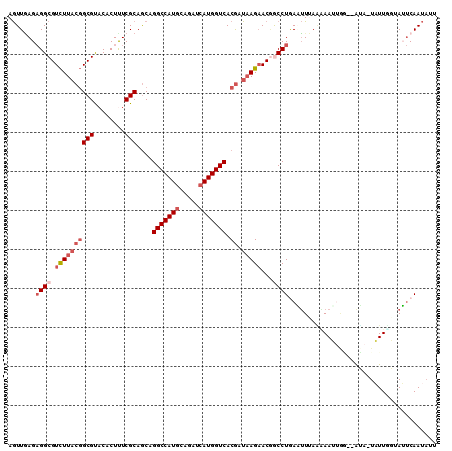
>2R_DroMel_CAF1 8391616 107 + 20766785 AGUUAAGAGGCGUCUUACGGCGUACACUUUCGCAGCAGGCCAUACAGAUCAUGGUCACGAUAAGAACGGCCUGAGUUUAAAAAUUGG--AUA-UAUUUGUAUUCAAUAUU .......((((.((((((((((........)))....((((((.......)))))).)).)))))...))))..........(((((--(((-(....)))))))))... ( -28.10) >DroGri_CAF1 6624 108 + 1 AAUUUAGCGGCGUUUUUCUGCGCACACUCUCGCAGCAGGCCAUGCAGAUCAUGGUCACAAUUAGCACGGCCUAAAAUCAGUAAAAAAAUACC-UAUU-UAAUACAACAGU ........((..((((((((((........))))).(((((.(((.(((..((....))))).))).)))))...........)))))..))-....-............ ( -23.00) >DroSec_CAF1 2732 107 + 1 AGUUGAGAGGCGUCUUACGGCGUACACUUUCGCAGCAGGCCAUGCAGAUCAUGGUCACGAUAAGAACGACCUAAGUUUAAGAAUUGG--AUA-UAUUUGCAUUCAAUAUU .(((((((((((((((((((((........)))....(((((((.....))))))).)).)))).))).)))..(((((.....)))--)).-........))))))... ( -27.20) >DroSim_CAF1 2740 109 + 1 AGUUGAGAGGCGUCUUACGGCGUACACUUUCGCAGCAGGCCAUGCAGAUCAUGGUCACGAUAAGAACGACCUGAAUUUAGGAAUAGGGAGUA-UAUUCGUAUUCAAUAUU .(((((((((((((((((((((........)))....(((((((.....))))))).)).)))).))).))).....((.(((((.......-))))).))))))))... ( -31.30) >DroEre_CAF1 4804 108 + 1 AGUUGAGAGGCGUCUUACGGCGUACACUCUCGCAACAGGCCAUGCAUAUCAUGGUCACGAUAAGAACAGCCUGAUUUGACAAAAUAU--AGAUUUUUGUUAAUCAAUAUA .(((((.((((.((((((((((........)))....(((((((.....))))))).)).)))))...))))...(((((((((...--....))))))))))))))... ( -34.70) >DroYak_CAF1 2730 108 + 1 AGUUGAGAGGCGUCUUACGGCGUACACUUUCGCAGCAGGCCAUGCAUAUCAUGGUCACGAUAAGAACUGCCUGAUUUUUACAAUUUC--GGAUUAUUGUUAAUCAAUUUA .......((((.((((((((((........)))....(((((((.....))))))).)).)))))...))))((((...(((((...--.....))))).))))...... ( -29.50) >consensus AGUUGAGAGGCGUCUUACGGCGUACACUUUCGCAGCAGGCCAUGCAGAUCAUGGUCACGAUAAGAACGGCCUGAAUUUAAAAAUUGG__AUA_UAUUGGUAUUCAAUAUU .......((((.((((((((((........)))....(((((((.....))))))).)).)))))...))))...................................... (-18.47 = -19.83 + 1.36)

| Location | 8,391,616 – 8,391,723 |
|---|---|
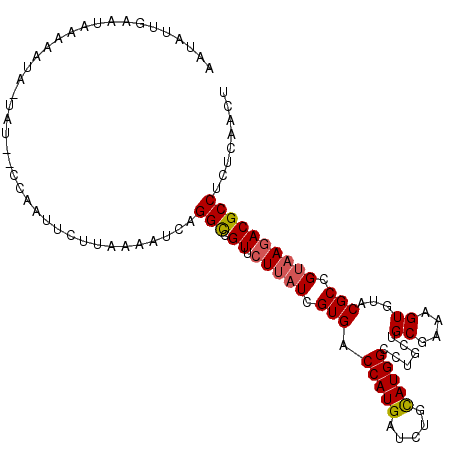
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.33 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -17.93 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
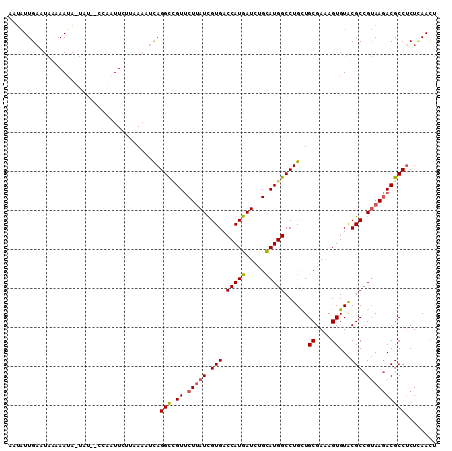
>2R_DroMel_CAF1 8391616 107 - 20766785 AAUAUUGAAUACAAAUA-UAU--CCAAUUUUUAAACUCAGGCCGUUCUUAUCGUGACCAUGAUCUGUAUGGCCUGCUGCGAAAGUGUACGCCGUAAGACGCCUCUUAACU .........(((.....-...--..............((((((((.(..(((((....)))))..).))))))))..((....))))).....(((((....)))))... ( -22.60) >DroGri_CAF1 6624 108 - 1 ACUGUUGUAUUA-AAUA-GGUAUUUUUUUACUGAUUUUAGGCCGUGCUAAUUGUGACCAUGAUCUGCAUGGCCUGCUGCGAGAGUGUGCGCAGAAAAACGCCGCUAAAUU (((.(((((..(-(((.-((((......)))).))))((((((((((..((..(....)..))..)))))))))).))))).)))(((.((........)))))...... ( -33.00) >DroSec_CAF1 2732 107 - 1 AAUAUUGAAUGCAAAUA-UAU--CCAAUUCUUAAACUUAGGUCGUUCUUAUCGUGACCAUGAUCUGCAUGGCCUGCUGCGAAAGUGUACGCCGUAAGACGCCUCUCAACU ....((((..((.....-...--................(((((.........)))))....(((..(((((.(((.((....))))).))))).))).))...)))).. ( -23.00) >DroSim_CAF1 2740 109 - 1 AAUAUUGAAUACGAAUA-UACUCCCUAUUCCUAAAUUCAGGUCGUUCUUAUCGUGACCAUGAUCUGCAUGGCCUGCUGCGAAAGUGUACGCCGUAAGACGCCUCUCAACU .....(((((..(((((-.......)))))....)))))((.(((.(((((.(((.(((((.....)))))..(((.((....)))))))).))))))))))........ ( -24.90) >DroEre_CAF1 4804 108 - 1 UAUAUUGAUUAACAAAAAUCU--AUAUUUUGUCAAAUCAGGCUGUUCUUAUCGUGACCAUGAUAUGCAUGGCCUGUUGCGAGAGUGUACGCCGUAAGACGCCUCUCAACU ......((((.((((((....--...))))))..))))(((((((.(.((((((....)))))).).)))))))((((.(((.((((.(.......)))))))).)))). ( -28.80) >DroYak_CAF1 2730 108 - 1 UAAAUUGAUUAACAAUAAUCC--GAAAUUGUAAAAAUCAGGCAGUUCUUAUCGUGACCAUGAUAUGCAUGGCCUGCUGCGAAAGUGUACGCCGUAAGACGCCUCUCAACU .....(((((.(((((.....--...)))))...)))))(((.((...((((((....)))))).))(((((.(((.((....))))).))))).....)))........ ( -26.60) >consensus AAUAUUGAAUAAAAAUA_UAU__CCAAUUCUUAAAAUCAGGCCGUUCUUAUCGUGACCAUGAUCUGCAUGGCCUGCUGCGAAAGUGUACGCCGUAAGACGCCUCUCAACU .......................................(((.((.(((((.(((.(((((.....)))))......((....))...))).))))))))))........ (-17.93 = -18.07 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:24 2006