
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,386,064 – 8,386,171 |
| Length | 107 |
| Max. P | 0.870218 |

| Location | 8,386,064 – 8,386,171 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.00 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
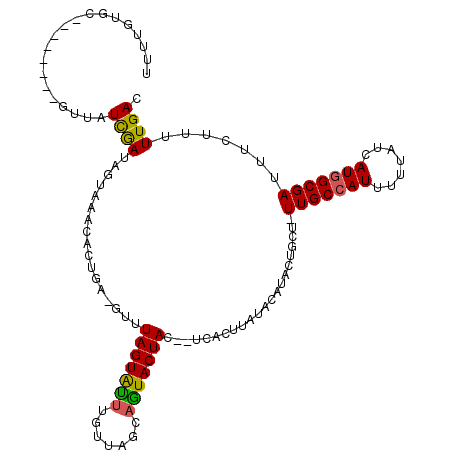
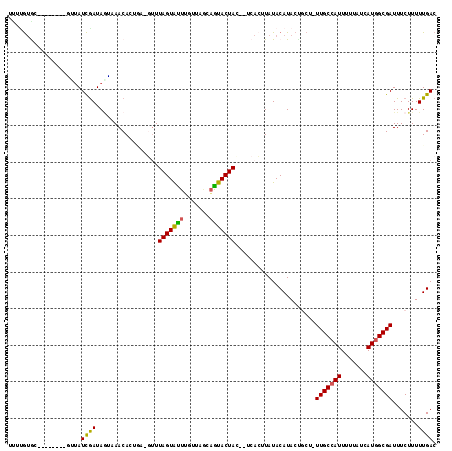
>2R_DroMel_CAF1 8386064 107 + 20766785 CCUUGUGA--------GCUAUCGAUAGUCAACACUCACGUUUAGUACUUCCUACCAGUACUAC--UCACAUAUACAUACACCUUUUGCCAUUUUUAUCAUGGCGAUUUCUUUUUGAC ...(((((--------(....((..(((....)))..))..(((((((.......))))))))--)))))....((........(((((((.......)))))))........)).. ( -20.09) >DroVir_CAF1 20531 107 + 1 UUUUGUGC------AUGUUAUCGAUAGCAAACACCGA-GUUUAGUGUUUGUUAACAAUACUAU--ACACUUAUACAUACUGCU-UUGCCAUUUUUAUCAUGGCGAUUUCUUUUUGAC ....(((.------(((.(((.(.((((((((((...-.....)))))))))).).))).)))--.)))..............-(((((((.......)))))))............ ( -22.30) >DroGri_CAF1 13804 109 + 1 UUUUGUAC------AAUGUAUCGAUAGAAAACACAAA-CCUUAGUGUUUGUUAGCAGUACUACGAACAGUUAUACAUACUGCU-UUGCCAUUUUUAUCAUGGCGAUUUCUUUUUGAC ........------......((((.(((((..(((((-(......)))))).(((((((..((.....))......)))))))-(((((((.......))))))))))))..)))). ( -22.10) >DroEre_CAF1 13549 107 + 1 CCUCGUGA--------GCUAUCGAUAGUCAACACUCACGUUUAGUACUGCCUAGCAGUACUAC--UCACAUAUACAUACCUCUUUUGCCAUUUUUAUCAUGGCGAUUUCUUUUUGAC ....((((--------(....((..(((....)))..))..(((((((((...))))))))))--)))).....((........(((((((.......)))))))........)).. ( -25.29) >DroWil_CAF1 30330 114 + 1 AAUUAUGGUCAUCG--AUUAUUAAUAAUCAACACUGAGUUUUAGUAUUGGUUAGACGUACUACCCUCACUUAUACAUACUGCU-UUGCAAUUUUUAUCAUGGCGAUUUCUUUUUGAC ....((.(((((.(--((......(((((((.(((((...))))).)))))))..........................(((.-..)))......)))))))).))........... ( -13.70) >DroMoj_CAF1 17443 107 + 1 UUAAGCAC------AUUUCAUCGAUAUGAAACACCGA-GUUUAGUGUUUGUUAACAGUACUAC--UCCCUUAUACAUACUGCU-UUGCCAUUUUUAUCAUGGCGAUUUCUUUUUGAC ...((((.------.((((((....))))))....((-((.((.((((....)))).))..))--))............))))-(((((((.......)))))))............ ( -19.30) >consensus UUUUGUGC________GUUAUCGAUAGUAAACACUGA_GUUUAGUAUUUGUUAGCAGUACUAC__UCACUUAUACAUACUGCU_UUGCCAUUUUUAUCAUGGCGAUUUCUUUUUGAC ....................((((.................(((((((.......)))))))......................(((((((.......))))))).......)))). (-10.55 = -10.00 + -0.55)

| Location | 8,386,064 – 8,386,171 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -10.60 |
| Energy contribution | -10.30 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
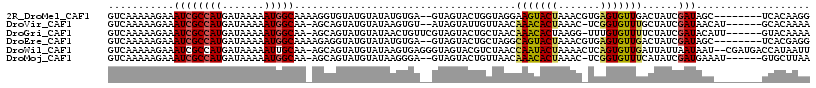
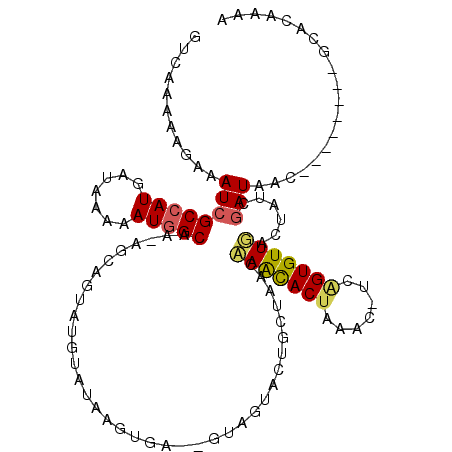
>2R_DroMel_CAF1 8386064 107 - 20766785 GUCAAAAAGAAAUCGCCAUGAUAAAAAUGGCAAAAGGUGUAUGUAUAUGUGA--GUAGUACUGGUAGGAAGUACUAAACGUGAGUGUUGACUAUCGAUAGC--------UCACAAGG ..............(((((.......))))).................((..--.(((((((.......))))))).))((((((((((.....))))).)--------)))).... ( -26.90) >DroVir_CAF1 20531 107 - 1 GUCAAAAAGAAAUCGCCAUGAUAAAAAUGGCAA-AGCAGUAUGUAUAAGUGU--AUAGUAUUGUUAACAAACACUAAAC-UCGGUGUUUGCUAUCGAUAACAU------GCACAAAA ..............(((((.......)))))..-..............((((--((..(((((.((.((((((((....-..)))))))).)).)))))..))------)))).... ( -28.40) >DroGri_CAF1 13804 109 - 1 GUCAAAAAGAAAUCGCCAUGAUAAAAAUGGCAA-AGCAGUAUGUAUAACUGUUCGUAGUACUGCUAACAAACACUAAGG-UUUGUGUUUUCUAUCGAUACAUU------GUACAAAA (((....(((((.((((((.......)))))..-((((((((.(((........))))))))))).((((((......)-)))))).)))))...))).....------........ ( -24.10) >DroEre_CAF1 13549 107 - 1 GUCAAAAAGAAAUCGCCAUGAUAAAAAUGGCAAAAGAGGUAUGUAUAUGUGA--GUAGUACUGCUAGGCAGUACUAAACGUGAGUGUUGACUAUCGAUAGC--------UCACGAGG ..............(((((.......))))).....................--.(((((((((...)))))))))..(((((((((((.....))))).)--------)))))... ( -31.70) >DroWil_CAF1 30330 114 - 1 GUCAAAAAGAAAUCGCCAUGAUAAAAAUUGCAA-AGCAGUAUGUAUAAGUGAGGGUAGUACGUCUAACCAAUACUAAAACUCAGUGUUGAUUAUUAAUAAU--CGAUGACCAUAAUU ((((.........(((....(((...(((((..-.)))))...)))...((((..(((((...........)))))...)))))))(((((((....))))--)))))))....... ( -17.10) >DroMoj_CAF1 17443 107 - 1 GUCAAAAAGAAAUCGCCAUGAUAAAAAUGGCAA-AGCAGUAUGUAUAAGGGA--GUAGUACUGUUAACAAACACUAAAC-UCGGUGUUUCAUAUCGAUGAAAU------GUGCUUAA ..............(((((.......))))).(-((((............((--((..((.((((....)))).)).))-))...(((((((....)))))))------.))))).. ( -21.50) >consensus GUCAAAAAGAAAUCGCCAUGAUAAAAAUGGCAA_AGCAGUAUGUAUAAGUGA__GUAGUACUGCUAACAAACACUAAAC_UCAGUGUUGACUAUCGAUAAC________GCACAAAA ...........((((((((.......))))).....................................(((((((.......)))))))......)))................... (-10.60 = -10.30 + -0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:20 2006