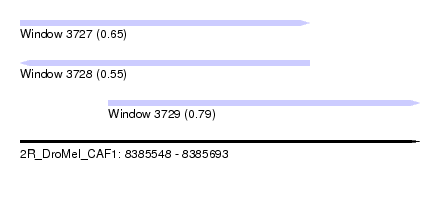
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,385,548 – 8,385,693 |
| Length | 145 |
| Max. P | 0.786744 |

| Location | 8,385,548 – 8,385,653 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -33.43 |
| Consensus MFE | -20.21 |
| Energy contribution | -22.77 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
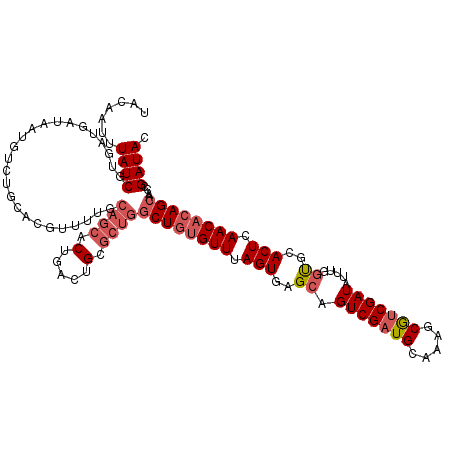
>2R_DroMel_CAF1 8385548 105 + 20766785 UACAAUUUAUCAUAAUGAUGA--------CGUUAUGCAGCACUGACUG------CUGAGUUUAGUGAGUAGUCGAUGCAAGCCAACGAUAGCUAUCGAACUCAACAAAGCAAUCGAUAC ..........(((((((....--------)))))))..(((.((((((------((.(......).)))))))).))).......((((.(((..............))).)))).... ( -24.34) >DroSec_CAF1 12482 119 + 1 UACAAUUUAUCGUGAUGAUAAUGUCUGCACGUUUUGCAGCACUGACUGCGCUGGCUGUGUUUAGUGAGCAGUCGAUGCAAAGCGUCGAUAUUUGGUGCACUCAACACAGCAACCGAUAC .......(((((((.......((.(((((.....))))))).......)))..((((((((.((((.((.((((((((...)))))))).....)).)))).))))))))....)))). ( -39.64) >DroSim_CAF1 12511 119 + 1 UACAAUUUAUCGUGAUGAUAAUGUCUGCACGCUUUACAGCACUGACUGCGCUGGCUGUGUUUAGUAAGCAGUCGAUGCAAAGCGUCGAUAUUUGGUGCACUCAACACAGCAACCGAUAC .......(((((((..(((...)))..)))......((((.(.....).))))((((((((.(((..((.((((((((...)))))))).....))..))).))))))))....)))). ( -36.30) >consensus UACAAUUUAUCGUGAUGAUAAUGUCUGCACGUUUUGCAGCACUGACUGCGCUGGCUGUGUUUAGUGAGCAGUCGAUGCAAAGCGUCGAUAUUUGGUGCACUCAACACAGCAACCGAUAC .......((((.........................((((.(.....).))))((((((((.(((..((.(((((((.....))))))).....))..))).))))))))....)))). (-20.21 = -22.77 + 2.56)

| Location | 8,385,548 – 8,385,653 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -19.29 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8385548 105 - 20766785 GUAUCGAUUGCUUUGUUGAGUUCGAUAGCUAUCGUUGGCUUGCAUCGACUACUCACUAAACUCAG------CAGUCAGUGCUGCAUAACG--------UCAUCAUUAUGAUAAAUUGUA ((((.(((((((..(((((((((((((((((....)))))...)))))..)))))....))..))------))))).))))((((....(--------((((....)))))....)))) ( -30.20) >DroSec_CAF1 12482 119 - 1 GUAUCGGUUGCUGUGUUGAGUGCACCAAAUAUCGACGCUUUGCAUCGACUGCUCACUAAACACAGCCAGCGCAGUCAGUGCUGCAAAACGUGCAGACAUUAUCAUCACGAUAAAUUGUA .((((((((((((((((.((((...((....((((.((...)).)))).))..)))).))))))).))))...(..(((((((((.....))))).))))..)....)))))....... ( -35.50) >DroSim_CAF1 12511 119 - 1 GUAUCGGUUGCUGUGUUGAGUGCACCAAAUAUCGACGCUUUGCAUCGACUGCUUACUAAACACAGCCAGCGCAGUCAGUGCUGUAAAGCGUGCAGACAUUAUCAUCACGAUAAAUUGUA (((.((.((((((((((.((((((.......((((.((...)).)))).)))..))).))))))))((((((.....))))))..)).)))))..((((((((.....)))))..))). ( -32.50) >consensus GUAUCGGUUGCUGUGUUGAGUGCACCAAAUAUCGACGCUUUGCAUCGACUGCUCACUAAACACAGCCAGCGCAGUCAGUGCUGCAAAACGUGCAGACAUUAUCAUCACGAUAAAUUGUA .(((((......(((((((............)))))))...((((.((((((..................)))))).))))..........................)))))....... (-19.29 = -19.40 + 0.12)

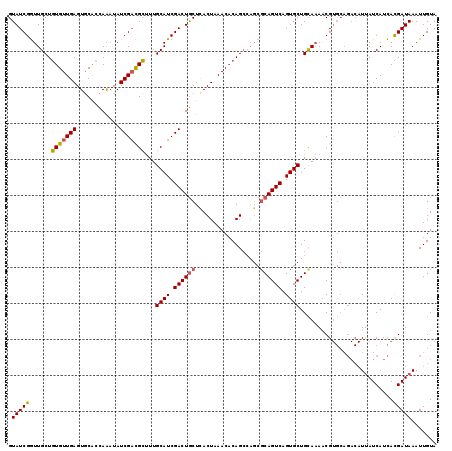
| Location | 8,385,580 – 8,385,693 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 87.11 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -21.43 |
| Energy contribution | -22.43 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
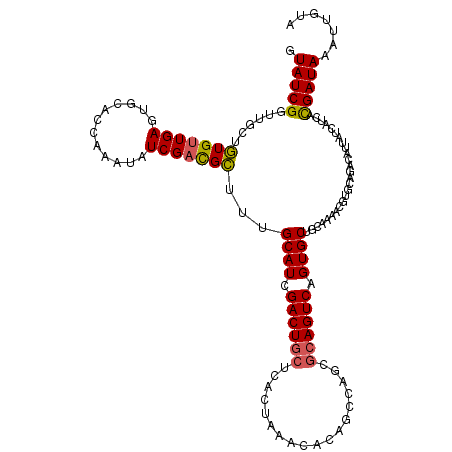
>2R_DroMel_CAF1 8385580 113 + 20766785 ACUGACUG------CUGAGUUUAGUGAGUAGUCGAUGCAAGCCAACGAUAGCUAUCGAACUCAACAAAGCAAUCGAUACUAUUGGAAUAGAAAAUAUAAGAAUUAAUUAAUUCAAACAU ..((((((------((.(......).))))))))...........((((((.((((((.((......))...))))))))))))...............(((((....)))))...... ( -21.90) >DroSec_CAF1 12522 119 + 1 ACUGACUGCGCUGGCUGUGUUUAGUGAGCAGUCGAUGCAAAGCGUCGAUAUUUGGUGCACUCAACACAGCAACCGAUACUAUUGGAAUAGAAAAAAUAAGAAUUAAUUAAUUCAAAUAU .(((.........((((((((.((((.((.((((((((...)))))))).....)).)))).))))))))..(((((...)))))..))).........(((((....)))))...... ( -33.60) >DroSim_CAF1 12551 119 + 1 ACUGACUGCGCUGGCUGUGUUUAGUAAGCAGUCGAUGCAAAGCGUCGAUAUUUGGUGCACUCAACACAGCAACCGAUACUAUUGGAAUAGAAAAAAUAAGAAUUAAUUAAUUCAAAUAU .(((.........((((((((.(((..((.((((((((...)))))))).....))..))).))))))))..(((((...)))))..))).........(((((....)))))...... ( -30.90) >consensus ACUGACUGCGCUGGCUGUGUUUAGUGAGCAGUCGAUGCAAAGCGUCGAUAUUUGGUGCACUCAACACAGCAACCGAUACUAUUGGAAUAGAAAAAAUAAGAAUUAAUUAAUUCAAAUAU .(((.........((((((((.(((..((.(((((((.....))))))).....))..))).))))))))..(((((...)))))..))).........(((((....)))))...... (-21.43 = -22.43 + 1.01)

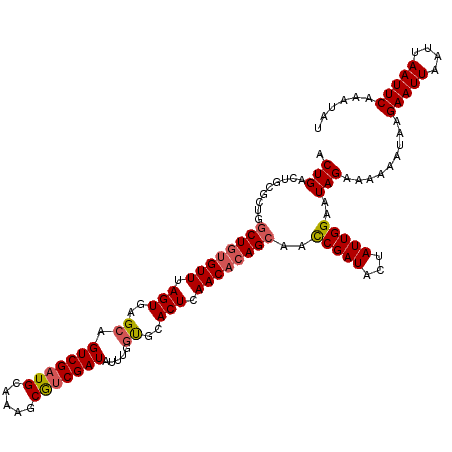
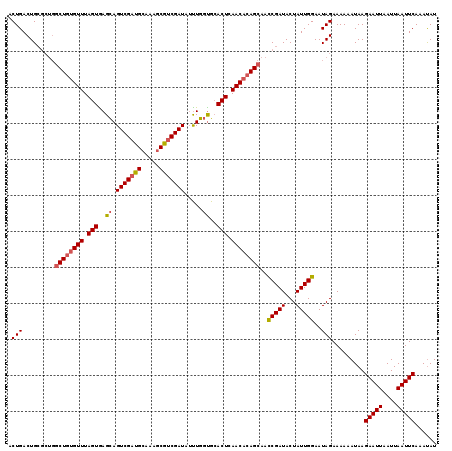
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:18 2006