| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,366,026 – 8,366,159 |
| Length | 133 |
| Max. P | 0.844171 |

| Location | 8,366,026 – 8,366,140 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -30.77 |
| Energy contribution | -30.25 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
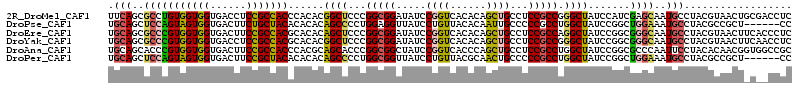
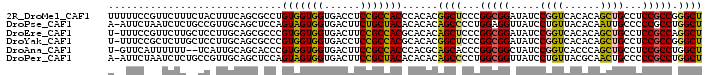
>2R_DroMel_CAF1 8366026 114 - 20766785 UUCAGCGCCUGUGGUGGUGACCUCCGCCACCCACACGGCUCCCGGCGGAUAUCCGGUCACACAGCUGCCUCCGCCGGGCUAUCCAUCGAGCAAUGCCUACGUAACUGCGACCUC ....(((((((((((((((.....))))).))))).))).(((((((((....((((......))))..)))))))))...........))........(((....)))..... ( -45.20) >DroPse_CAF1 4716 108 - 1 UGCAGCUCCAGUAGUGGUGACUUCUGCUACACACACAGCCCCUGGAGGUUAUCCUGUUACACAAUUGCCCCCGCCUGGCUAUCCGGCUGGAAAUGCCUACGCCGCU------CC .(((..((((((.((((((..........))).)))((((..(((.(((.....((.....))...))).)))...)))).....))))))..)))..........------.. ( -28.80) >DroEre_CAF1 4616 114 - 1 UGCAGCGCCCGUGGUGGUGACUUCCGCCACGCACACAGCUCCCGGCGGAUAUCCGGUCACACAGCUGCCUCCGCCAGGCUAUCCGGCGGGCAAUGCCUACGUAACUUCACCCUC .(((..(((((((((((......)))))))((....((((...((((((....((((......))))..)))))).)))).....))))))..))).................. ( -44.50) >DroYak_CAF1 4633 114 - 1 UGCAGCGCCCGUGGUGGUGACCUCCGCCACGCACACGGCUCCCGGCGGAUAUCCGGUCACACAGCUGCCUCCGCCGGGCUAUCCGGCGGGCAAUGCCUACGUAACUUCAACCUC .(((..(((((((((((......)))))))((...(((..(((((((((....((((......))))..)))))))))....)))))))))..))).................. ( -51.30) >DroAna_CAF1 14631 114 - 1 UGCAGCACCCGUGGUGGUGACUUCCGCCACCCACGCAGCACCCGGCGGCUAUCCGGUCACCCAGCUGCCUCCGCCUGGCUAUCCGGCGCCCAAUUCCUACACAACGGUGGCCGC .((.((...((((((((((.....))))).)))))...((((.(((((((....((....)))))))))..((((.((....)))))).................)))))).)) ( -41.80) >DroPer_CAF1 4688 108 - 1 UGCAGCUCCAGUAGUGGUGACUUCCGCUACACACACAGCCCCUGGCGGUUAUCCUGUUACGCAACUGCCCCCGCCUGGCUAUCCGGCUGGAAAUGCCUACGCCGCU------CC .(((..((((((.((((((.....))))))......((((...(((((.......(((....))).....))))).)))).....))))))..)))..........------.. ( -34.80) >consensus UGCAGCGCCCGUGGUGGUGACUUCCGCCACACACACAGCUCCCGGCGGAUAUCCGGUCACACAGCUGCCUCCGCCUGGCUAUCCGGCGGGCAAUGCCUACGCAACU_C__CCUC .(((..(((((((((((......)))))))......((((...(((((.....((((......))))...))))).)))).......))))..))).................. (-30.77 = -30.25 + -0.52)
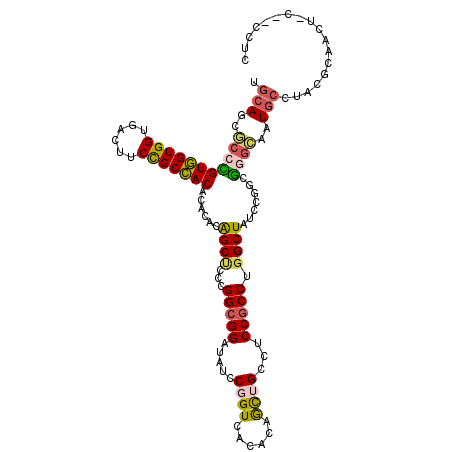
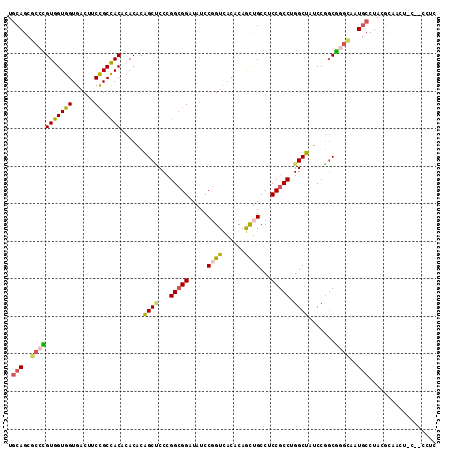
| Location | 8,366,060 – 8,366,159 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.41 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -23.69 |
| Energy contribution | -24.33 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8366060 99 + 20766785 AGCCCGGCGGAGGCAGCUGUGUGACCGGAUAUCCGCCGGGAGCCGUGUGGGUGGCGGAGGUCACCACCACAGGCGCUGAAAGUAGAAAGAACGGAAAAA ..(((((((((.....(((......)))...))))))))).((((((..((((((....))))))..))).)))......................... ( -41.30) >DroPse_CAF1 4744 98 + 1 AGCCAGGCGGGGGCAAUUGUGUAACAGGAUAACCUCCAGGGGCUGUGUGUGUAGCAGAAGUCACCACUACUGGAGCUGCAACGGCAGAGAUUAGAAU-U ..((((..(((((..(((.((...)).)))..))))).((((((.(((.....)))..)))).))....))))..((((....))))..........-. ( -28.50) >DroEre_CAF1 4650 98 + 1 AGCCUGGCGGAGGCAGCUGUGUGACCGGAUAUCCGCCGGGAGCUGUGUGCGUGGCGGAAGUCACCACCACGGGCGCUGCAAGGAGCAAGAACGGAAA-A ..(((((((((.....(((......)))...))))))))).((.((((.(((((.((......)).))))).)))).))..................-. ( -41.90) >DroYak_CAF1 4667 98 + 1 AGCCCGGCGGAGGCAGCUGUGUGACCGGAUAUCCGCCGGGAGCCGUGUGCGUGGCGGAGGUCACCACCACGGGCGCUGCAAGGAGCAAGAGCGGAAA-A ..(((((((((.....(((......)))...))))))))).((.((((.(((((.((......)).))))).)))).)).....((....)).....-. ( -45.30) >DroAna_CAF1 14665 96 + 1 AGCCAGGCGGAGGCAGCUGGGUGACCGGAUAGCCGCCGGGUGCUGCGUGGGUGGCGGAAGUCACCACCACGGGUGCUGCAAUGA--AAAAAAUGAAC-A .((..(((((......((((....))))....))))).((..((.((((((((((....)))))..)))))))..)))).....--...........-. ( -43.60) >DroPer_CAF1 4716 98 + 1 AGCCAGGCGGGGGCAGUUGCGUAACAGGAUAACCGCCAGGGGCUGUGUGUGUAGCGGAAGUCACCACUACUGGAGCUGCAACGGCAGAGAUUAGAAU-U .(((..((....)).(((((((..(((....((.(((...))).))(((.((..(....)..)))))..)))..)).))))))))............-. ( -31.10) >consensus AGCCAGGCGGAGGCAGCUGUGUGACCGGAUAUCCGCCGGGAGCUGUGUGCGUGGCGGAAGUCACCACCACGGGCGCUGCAAGGAGAAAGAACGGAAA_A .(((((((((..((......))..((((.......))))...))))(((.(((((....))))))))..)))))......................... (-23.69 = -24.33 + 0.64)


| Location | 8,366,060 – 8,366,159 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.41 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -24.08 |
| Energy contribution | -23.42 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8366060 99 - 20766785 UUUUUCCGUUCUUUCUACUUUCAGCGCCUGUGGUGGUGACCUCCGCCACCCACACGGCUCCCGGCGGAUAUCCGGUCACACAGCUGCCUCCGCCGGGCU ......................((((((((((((((((.....))))).))))).)))..((((((((....((((......))))..))))))))))) ( -41.40) >DroPse_CAF1 4744 98 - 1 A-AUUCUAAUCUCUGCCGUUGCAGCUCCAGUAGUGGUGACUUCUGCUACACACACAGCCCCUGGAGGUUAUCCUGUUACACAAUUGCCCCCGCCUGGCU .-..........((((....)))).....((((..(......)..))))......((((..(((.(((.....((.....))...))).)))...)))) ( -21.50) >DroEre_CAF1 4650 98 - 1 U-UUUCCGUUCUUGCUCCUUGCAGCGCCCGUGGUGGUGACUUCCGCCACGCACACAGCUCCCGGCGGAUAUCCGGUCACACAGCUGCCUCCGCCAGGCU .-.........((((.....)))).(((((((((((......))))))))............((((((....((((......))))..)))))).))). ( -35.10) >DroYak_CAF1 4667 98 - 1 U-UUUCCGCUCUUGCUCCUUGCAGCGCCCGUGGUGGUGACCUCCGCCACGCACACGGCUCCCGGCGGAUAUCCGGUCACACAGCUGCCUCCGCCGGGCU .-.....(((...((.....)))))(((((((((((......)))))))).....))).(((((((((....((((......))))..))))))))).. ( -42.50) >DroAna_CAF1 14665 96 - 1 U-GUUCAUUUUUU--UCAUUGCAGCACCCGUGGUGGUGACUUCCGCCACCCACGCAGCACCCGGCGGCUAUCCGGUCACCCAGCUGCCUCCGCCUGGCU .-...........--.....((.((....(((((((((.....))))).))))(((((....((.((((....)))).))..)))))....))...)). ( -30.60) >DroPer_CAF1 4716 98 - 1 A-AUUCUAAUCUCUGCCGUUGCAGCUCCAGUAGUGGUGACUUCCGCUACACACACAGCCCCUGGCGGUUAUCCUGUUACGCAACUGCCCCCGCCUGGCU .-..........((((....)))).....(((((((......)))))))......((((...(((((.......(((....))).....))))).)))) ( -27.90) >consensus U_UUUCCGUUCUUUCUCCUUGCAGCGCCCGUGGUGGUGACUUCCGCCACACACACAGCUCCCGGCGGAUAUCCGGUCACACAGCUGCCUCCGCCUGGCU .............................(((((((......)))))))......((((...(((((.....((((......))))...))))).)))) (-24.08 = -23.42 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:15 2006