
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,360,331 – 8,360,427 |
| Length | 96 |
| Max. P | 0.516749 |

| Location | 8,360,331 – 8,360,427 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 65.21 |
| Mean single sequence MFE | -13.98 |
| Consensus MFE | -8.96 |
| Energy contribution | -8.68 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8360331 96 + 20766785 AUUCCUCUACUUAUCCGCUGG---------------UAA--AAUUAAUUUAAUUUCAGAUACUACUGGGCAAUAUACGCCCCAUCACUUUGGAACUGUUCCAAAACCUACUGA .......((((........))---------------)).--.............((((........((((.......))))......((((((.....)))))).....)))) ( -15.60) >DroVir_CAF1 84198 87 + 1 A---------------UUGAAU--------U-AACUUGAAUACUU--GACUAAUGCAGAUACUCCUCGGCAAUAUACGACCCAUUACAUUGGAGCUAUUUCAGAAUUUACUAA .---------------.(((((--------(-...(((((.....--......(((.((......)).)))......(..(((......)))..)...))))))))))).... ( -8.70) >DroSec_CAF1 71980 79 + 1 ----------------AUGCG-------------UUUGA--AAU---UUAAAUUCCAGAUACUACUGGGCAAUAUACGUCCCAUCACCUUGGAACUGUUCCAGAAUCUACUGA ----------------.((((-------------(((((--...---)))))).((((......)))))))......((.(((......))).)).....(((......))). ( -12.00) >DroSim_CAF1 71199 95 + 1 AUUCCUCUACUUAUUAAUCAG-------------UUUGA--AAU---UUAAAUUCCAGAUACUACUGGGCAAUAUACGUCCCAUCACCUUGGAACUGUUCCAAAAUCUACUGA .................((((-------------(..((--...---......(((((......)))))...................(((((.....)))))..)).))))) ( -13.70) >DroEre_CAF1 76592 88 + 1 UCUUA----AUUAUCCGCUGGUAA------------------UU---UAAAAUUCCAGAUACUCCUGGGCAACAUACGCCCCAUUACCUUGGAGCUUUUCCAAAAUCUACUGA .....----....((((..(((((------------------(.---.........((......))((((.......)))).)))))).)))).................... ( -14.80) >DroYak_CAF1 70102 104 + 1 UUUCC----CCUUUCCGCUGGUAAAACCUAUUACAUUGA--GCU---UCAAAUUACAGAUACUUCUGGGCAACAUACGCCCCAUUACUUUGGAGCUUUUCCAAAAUCUACUGA (((..----((........))..)))...........((--(((---(((((...((((....))))(((.......))).......))))))))))................ ( -19.10) >consensus AUUCC____CUUAUCCGCUGG______________UUGA__AAU___UUAAAUUCCAGAUACUACUGGGCAAUAUACGCCCCAUCACCUUGGAACUGUUCCAAAAUCUACUGA .......................................................(((........((((.......)))).......((((((...))))))......))). ( -8.96 = -8.68 + -0.28)

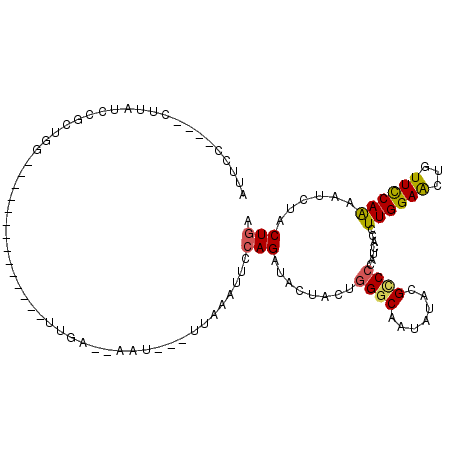
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:11 2006