| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,286,702 – 8,286,876 |
| Length | 174 |
| Max. P | 0.690038 |

| Location | 8,286,702 – 8,286,822 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.66 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
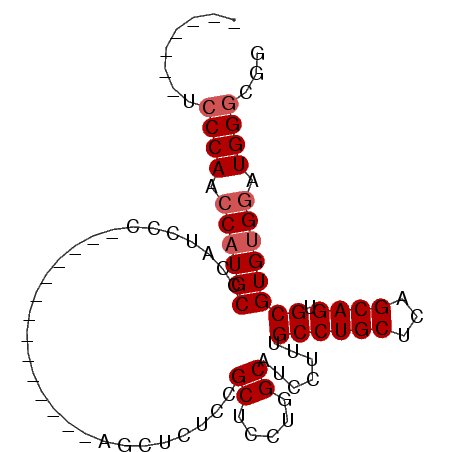
>2R_DroMel_CAF1 8286702 120 + 20766785 UAUUUCAUUCGCAAUGACGCUGACAGGACAAUGCGGUCCUGCAUGUCCUGGCUGCCGGCACGCCGUCUGCGCCAUCCAUCGUUCCCAUCCCAUCCCUGCACAUCCCAACCCUUCCCCUAC ..........((((((.(((.(((((((((.((((....))))))))))(((((....)).)))))).))).))).....................)))..................... ( -28.89) >DroSec_CAF1 9730 97 + 1 UAUUUCAUUCGCAAUGACGCUGACAGGACAAUGCGGUCCUGCAUGUCCUGGCUGCCGGCUCGCCGCCUCCGCCAUCCA---------UCCCAACCAUGCCGAUCCC-------------- ........(((((..((((.((.((((((......)))))))))))).((((.(.(((....))).)...))))....---------.........)).)))....-------------- ( -26.10) >DroSim_CAF1 8071 97 + 1 UAUUUCAUUCGCAAUGACGCUGACAGGACAAUGCGGUCCUGCAUGUCCUGGCUGCCGGCUCGCCGCCUCCGCCAUCCA---------UCCCAACCAUGCCCAUCCC-------------- ..........(((..((((.((.((((((......)))))))))))).((((.(.(((....))).)...))))....---------.........))).......-------------- ( -25.90) >DroYak_CAF1 8716 96 + 1 UAUUUCACUCGCAAUGACGCUGACAGGACAAUGCGGUCCUGCAUGUCCUGGCUGCCGGCUCGCCGCCUCCGUCAUCCA---------UCCUUCCCAUUCCC-UUCC-------------- .............(((((((...(((((((.((((....)))))))))))...)).(((.....)))...)))))...---------..............-....-------------- ( -27.30) >DroAna_CAF1 8468 94 + 1 UAUUUCAUUCGCAAUGACGCUGACAGGACAAUGCGGUCCUGCAUGUCCUGGCUGCUGCUUCACCGCC--CAUAGACCA---------CCCUCCCCUCUCCC-ACUC-------------- ..........((..(((.((.(.(((((((.((((....)))))))))))....).)).)))..)).--.........---------..............-....-------------- ( -21.60) >consensus UAUUUCAUUCGCAAUGACGCUGACAGGACAAUGCGGUCCUGCAUGUCCUGGCUGCCGGCUCGCCGCCUCCGCCAUCCA_________UCCCAACCAUGCCCAUCCC______________ ..........((......((...(((((((.((((....)))))))))))...)).((....))))...................................................... (-20.62 = -20.66 + 0.04)


| Location | 8,286,742 – 8,286,851 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.28 |
| Mean single sequence MFE | -18.23 |
| Consensus MFE | -16.70 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | 0.04 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8286742 109 + 20766785 GCAUGUCCUGGCUGCCGGCACGCCGUCUGCGCCAUCCAUCGUUCCCAUCCCAUCCCUGCACAUCCCAACCCUUCCCCUACAGCUGUCCGCUCCUGGCAUCCUUUGCCUG (((((...((((.((.(((.....))).))))))..))).))......................................(((.....)))...((((.....)))).. ( -17.60) >DroSec_CAF1 9770 86 + 1 GCAUGUCCUGGCUGCCGGCUCGCCGCCUCCGCCAUCCA---------UCCCAACCAUGCCGAUCCC--------------AGCUUUCCGCUCCUGGCAUCCUUUGCCUG (((((...((((.(.(((....))).)...))))....---------.......))))).......--------------(((.....)))...((((.....)))).. ( -18.54) >DroSim_CAF1 8111 86 + 1 GCAUGUCCUGGCUGCCGGCUCGCCGCCUCCGCCAUCCA---------UCCCAACCAUGCCCAUCCC--------------AGCUCUCCGCUCCUGGCAUCCUUUGCCUG (((((...((((.(.(((....))).)...))))....---------.......))))).......--------------(((.....)))...((((.....)))).. ( -18.54) >consensus GCAUGUCCUGGCUGCCGGCUCGCCGCCUCCGCCAUCCA_________UCCCAACCAUGCCCAUCCC______________AGCUCUCCGCUCCUGGCAUCCUUUGCCUG (((((...((((.(.(((....))).)...))))....................))))).....................(((.....)))...((((.....)))).. (-16.70 = -17.03 + 0.33)

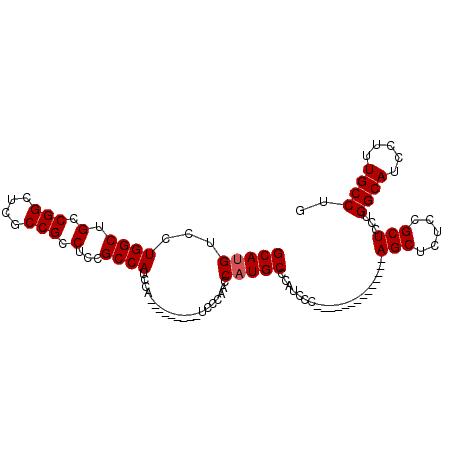
| Location | 8,286,782 – 8,286,876 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -17.10 |
| Energy contribution | -18.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8286782 94 + 20766785 GUUCCCAUCCCAUCCCUGCACAUCCCAACCCUUCCCCUACAGCUGUCCGCUCCUGGCAUCCUUUGCCUGCUCAGCAGUGCGUGUGGAUGGUCGG ....((...((((((((((((..........(......)..((((...((....((((.....)))).)).)))).))))).).))))))..)) ( -26.50) >DroSec_CAF1 9808 73 + 1 -------UCCCAACCAUGCCGAUCCC--------------AGCUUUCCGCUCCUGGCAUCCUUUGCCUGCUCAGCAGUGCGUGUGUAUGGGCGG -------..((..((((((((((.((--------------((..........)))).)))....((((((...)))).))..).))))))..)) ( -21.80) >DroSim_CAF1 8149 73 + 1 -------UCCCAACCAUGCCCAUCCC--------------AGCUCUCCGCUCCUGGCAUCCUUUGCCUGCUCAGCAGUGCGUGUGGAUGGGCGG -------.........((((((((((--------------(((.((..(((...((((.....)))).....))))).)).)).))))))))). ( -29.80) >consensus _______UCCCAACCAUGCCCAUCCC______________AGCUCUCCGCUCCUGGCAUCCUUUGCCUGCUCAGCAGUGCGUGUGGAUGGGCGG ........((((.((((((.............................((.....)).......((((((...)))).)))))))).))))... (-17.10 = -18.10 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:48 2006