| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,284,857 – 8,284,970 |
| Length | 113 |
| Max. P | 0.609130 |

| Location | 8,284,857 – 8,284,970 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.80 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -17.48 |
| Energy contribution | -18.65 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
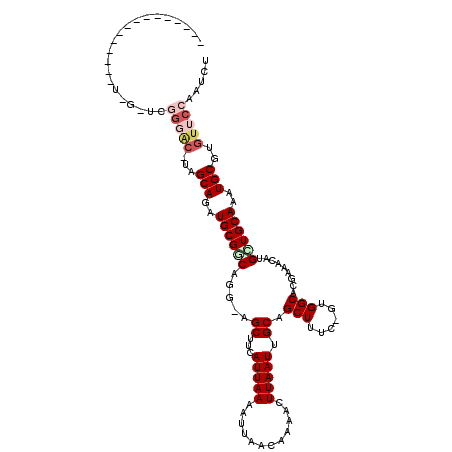
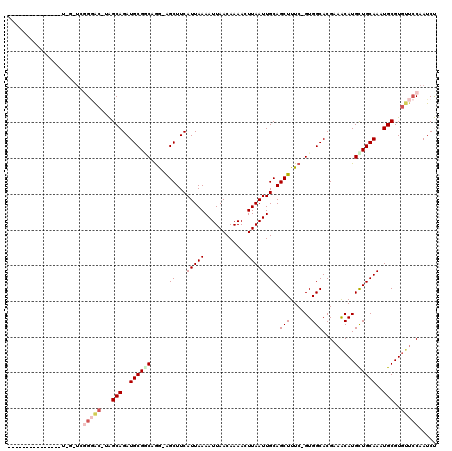
>2R_DroMel_CAF1 8284857 113 - 20766785 -----UUUUUGUUGGUCGAUCGGGGC-AAGCAGAUGCGCCUGGUGGCUUCAUUAAAAUUAACAAAACUUAAUUGCAGCUUUC-GUGGCACGAAACAUGCUGCAAAUGCGUGUUCCAAUCU -----............(((.(((((-(.(((((((.(((....)))..))))..................(((((((((((-(.....)))))...))))))).))).)))))).))). ( -33.90) >DroPse_CAF1 12137 98 - 1 -------------------UUGGGAC-UGGCAGAUGCGGCAGG-AGCUUCAUUAAAAUUAACAAAACUUAAUUGCAGCUUUC-GUGGCACGUAGCAUGUUGCAAAUGCGUGUUCCAAUCC -------------------(((((((-..(((..(((((((..-.(((.(.....((((((......))))))...(((...-..)))..).))).)))))))..)))..)))))))... ( -28.70) >DroWil_CAF1 12430 115 - 1 UUUUCCUUUUGCU-GUUGUU--GAAG-GAGCAGAUGCGACAAA-AGCUUCAUUAAAAUUAACAAAAUUUAAUUGCAGCUUUCGGUGGCACGUAGCAUGUUGCAAAUGCGUAGGGCAAUCU ...((((((.((.-...)).--))))-))(((..(((((((((-((((.((...(((((.....)))))...)).)))))).....((.....)).)))))))..)))............ ( -29.40) >DroYak_CAF1 6920 104 - 1 --------------UUUGGACGGGGC-AAGCAGAUGCGCCUGGUGGCUUCAUUAAAAUUAACAAAACUUAAUUGCAGCUUUC-GUGGCACGAAACAUGCUGCAAAUGCGUGUUCCAAUCU --------------.......(((((-(.(((((((.(((....)))..))))..................(((((((((((-(.....)))))...))))))).))).))))))..... ( -32.00) >DroAna_CAF1 6741 113 - 1 -----CAUUUAUUUAUAUAUAUGUGCAUAGCAGAUGCGGCUGG-GGCUUCAUUAAAAUUAACAAAACUUAAUUGCAGCUUUC-GUGGCACGAAACAUGCUGCAAAUGCGUGUUCCAAUCU -----...................((((.....))))(..(((-(((..((((..((((((......))))))(((((((((-(.....)))))...))))).))))...))))))..). ( -26.20) >DroPer_CAF1 10761 98 - 1 -------------------UUGGGAC-UGGCAGAUGCGGCAGG-AGCUUCAUUAAAAUUAACAAAACUUAAUUGCAGCUUUC-GUGGCACGUAGCAUGUUGCAAAUGCGUGUUCCAAUCC -------------------(((((((-..(((..(((((((..-.(((.(.....((((((......))))))...(((...-..)))..).))).)))))))..)))..)))))))... ( -28.70) >consensus _______________U_G_UCGGGAC_UAGCAGAUGCGGCAGG_AGCUUCAUUAAAAUUAACAAAACUUAAUUGCAGCUUUC_GUGGCACGAAACAUGCUGCAAAUGCGUGUUCCAAUCU .....................(((((...(((..((((((.....((...(((((............))))).)).(((......))).........))))))..)))..)))))..... (-17.48 = -18.65 + 1.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:45 2006