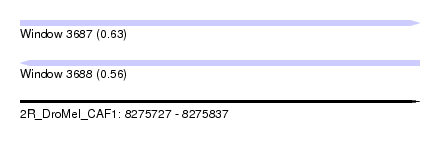
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,275,727 – 8,275,837 |
| Length | 110 |
| Max. P | 0.633358 |

| Location | 8,275,727 – 8,275,837 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.67 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -16.44 |
| Energy contribution | -16.67 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8275727 110 + 20766785 AAACAACAAAAUCAGG-AGUGCAGCUAACGCUGAAGCCCGCUCUAAAGCAUAGUAAGCUCCAUGAUGGCGUUGACGAUGUCAUUGUUGUUGUUCUUCAGAGCCUUGAUCGC .((((((((.((((((-(((.((((....))))......(((....))).......))))).))))((((((...)))))).))))))))((((....))))......... ( -33.20) >DroVir_CAF1 2323 103 + 1 AAUCAACGAAAGCAGC-GAUGUUCC----UCCGUG---GCCCUUAGAGCAUAGUAAGCUCCAUAAUGGCAUUAACGAUGUCGUUGUUGUUGUUUUUCAGUGCCUUUAUGGC ....(((((.((((((-(((((...----..(((.---(((.(((((((.......))))..))).)))....)))))))))))))).))))).......(((.....))) ( -29.00) >DroPse_CAF1 615 110 + 1 AAGCAACGAAAUCAUA-GUUUCUGCGGCGCGUGGAGAGCUAUCUAGAGCAUGGUGAGCUCCAUGAUGGCAUUGACUAUGUCGUUGCUGUUGUUCUUCAGCGCCUUGAUGGC .((((((((...((((-(((..(((.(..(((((((.((((((....).)))))...))))))).).)))..)))))))))))))))((((.....))))(((.....))) ( -44.00) >DroGri_CAF1 2234 105 + 1 GUUCAACGAAAGCAAC-GACGUUCC---CUGCG--CAUCGCCUUAGAGCAUGGUCAGCUCCAUGAUGGCGUUAACGAUAUCGUUGUUGUUGUUCUUCAGCGCCUUGAUGGC (((.((.(((((((((-((((....---...((--...((((......(((((......)))))..))))....))....)))))))))).))))).)))(((.....))) ( -31.10) >DroAna_CAF1 2116 108 + 1 AAACAACGAAAGCAGUGGACGCAGCAGCUACUGAAGCGC---CUAGAGCAUAGUCAGCUCCAUGAUGGCGUUAACUAUGUCGUUGCUGUUGUUCUUCAGCGCCUUGAUGGC ...........((...((((((((((((.((...(((((---(..((((.......))))......))))))......)).))))))).)))))....))(((.....))) ( -33.90) >DroPer_CAF1 2186 110 + 1 AAGCAACGAAAUCAUA-GUUUCUGCGGCGCGUGGAGAGCUAUCUAGAGCAUGGUGAGCUCCAUGAUGGCAUUGACUAUGUCGUUGCUGUUGUUCUUCAGCGCCUUGAUGGC .((((((((...((((-(((..(((.(..(((((((.((((((....).)))))...))))))).).)))..)))))))))))))))((((.....))))(((.....))) ( -44.00) >consensus AAACAACGAAAGCAGA_GAUGCUGC_GCCCCUGAAGAGCGCUCUAGAGCAUAGUAAGCUCCAUGAUGGCAUUAACGAUGUCGUUGCUGUUGUUCUUCAGCGCCUUGAUGGC .(((((((.....................................((((.......))))...((((((((.....))))))))..))))))).......(((.....))) (-16.44 = -16.67 + 0.22)



| Location | 8,275,727 – 8,275,837 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.67 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8275727 110 - 20766785 GCGAUCAAGGCUCUGAAGAACAACAACAAUGACAUCGUCAACGCCAUCAUGGAGCUUACUAUGCUUUAGAGCGGGCUUCAGCGUUAGCUGCACU-CCUGAUUUUGUUGUUU ((((.(((((((((((((...........((((...)))).......(((((......))))))))))))))(((...((((....))))...)-))....)))))))).. ( -28.30) >DroVir_CAF1 2323 103 - 1 GCCAUAAAGGCACUGAAAAACAACAACAACGACAUCGUUAAUGCCAUUAUGGAGCUUACUAUGCUCUAAGGGC---CACGGA----GGAACAUC-GCUGCUUUCGUUGAUU (((.....)))............((((((((....))))...(((....((((((.......))))))..)))---...(((----((......-....)))))))))... ( -24.90) >DroPse_CAF1 615 110 - 1 GCCAUCAAGGCGCUGAAGAACAACAGCAACGACAUAGUCAAUGCCAUCAUGGAGCUCACCAUGCUCUAGAUAGCUCUCCACGCGCCGCAGAAAC-UAUGAUUUCGUUGCUU (((.....))).............((((((((((((((...(((.(((.((((((.......))))))))).((.......))...)))...))-))))...)))))))). ( -34.70) >DroGri_CAF1 2234 105 - 1 GCCAUCAAGGCGCUGAAGAACAACAACAACGAUAUCGUUAACGCCAUCAUGGAGCUGACCAUGCUCUAAGGCGAUG--CGCAG---GGAACGUC-GUUGCUUUCGUUGAAC (((.....)))...............((((((....((...((((....((((((.......)))))).))))..)--)((((---.(.....)-.))))..))))))... ( -31.10) >DroAna_CAF1 2116 108 - 1 GCCAUCAAGGCGCUGAAGAACAACAGCAACGACAUAGUUAACGCCAUCAUGGAGCUGACUAUGCUCUAG---GCGCUUCAGUAGCUGCUGCGUCCACUGCUUUCGUUGUUU (((.....)))((((........))))(((((..((((..((((.....((((((.......))))))(---(((((.....))).)))))))..))))...))))).... ( -34.30) >DroPer_CAF1 2186 110 - 1 GCCAUCAAGGCGCUGAAGAACAACAGCAACGACAUAGUCAAUGCCAUCAUGGAGCUCACCAUGCUCUAGAUAGCUCUCCACGCGCCGCAGAAAC-UAUGAUUUCGUUGCUU (((.....))).............((((((((((((((...(((.(((.((((((.......))))))))).((.......))...)))...))-))))...)))))))). ( -34.70) >consensus GCCAUCAAGGCGCUGAAGAACAACAACAACGACAUAGUCAACGCCAUCAUGGAGCUCACCAUGCUCUAGAGAGCUCUCCACACGC_GCAGCAUC_GCUGAUUUCGUUGAUU (((.....))).............((((((((....((....)).....((((((.......))))))..................................)))))))). (-16.29 = -16.35 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:39 2006