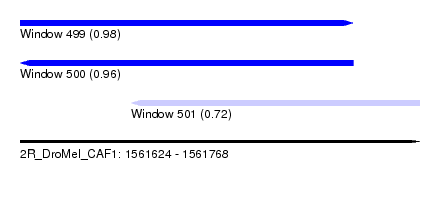
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,561,624 – 1,561,768 |
| Length | 144 |
| Max. P | 0.976197 |

| Location | 1,561,624 – 1,561,744 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.75 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -33.74 |
| Energy contribution | -34.78 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1561624 120 + 20766785 GAUAUAUAAAACUGAUUAAUGUCGGCACUGGUUAUUAAUGGCCAUAAGGCUGCAGAGGCCCAGAGCACAGGCUCAUGCUCUGAUUGCGGUGAAGAUGAGCUUGGGCGAGCAGCAUAAUUA ...........(((((....)))))...((((((....))))))....(((((....(((((((((....))))..((((...((......))...)))).)))))..)))))....... ( -35.40) >DroSec_CAF1 11365 120 + 1 GAUAUAUAAAACUGAUUAAUGUCGGCACUGGUUAUUAAUGGCCAUAAGGCUGCAGAGGCCCAGUGCUCAUUCUCAUGCUCUGAUUGCGGUGAAGAUGAGCUCGGGCGAGCAGCAUAAUUA ...........(((((....)))))...((((((....))))))....(((((....((((.(.((((((..((((((.......)).))))..)))))).)))))..)))))....... ( -40.60) >DroSim_CAF1 12898 120 + 1 GAUAUAUAAAACUGAUUAAUGUCGGCACUGGUUAUUAAUGGCCAUAAGGCUGCAGAGGCCCAGUGCUCAGGCUCAUGCUCUGAUUGCGGUGAAGAUGAGCUCGGGCGAGCAGCAUAAUUA ...........(((((....)))))...((((((....))))))....(((((....((((.(.(((((...((((((.......)).))))...))))).)))))..)))))....... ( -37.00) >DroEre_CAF1 11740 120 + 1 GAUAUAUAAAACUGAUUAAUGUCGGCGCUGGUUAUUAAUGGCCAUAAGGCUGCGGAGGCCCAGUGCCCAGGCUCAUGCUCUGAUUGCGCCGAAGAUGAGCUUGGGCGAGCAGCAUAAUUA ...........(((((....)))))...((((((....))))))....(((((((....))..(((((((((((((.((.((.......)).))))))))))))))).)))))....... ( -42.70) >DroYak_CAF1 17728 120 + 1 GAUAUAUAAAACUGAUUAAUGUCGGCGCUGGUUAUUAAUGACCAUAAGGCAGCAGAGGCCCAGUGCUCAGGCUCAUGCUCUGAUUGCGGUGAAGAUGAGCUUGGGCGAGCAGCAUAAUUA ...........(((((....))))).((((.((..............(((.......)))....((((((((((((.(.(((....))).)...)))))))))))))).))))....... ( -36.20) >consensus GAUAUAUAAAACUGAUUAAUGUCGGCACUGGUUAUUAAUGGCCAUAAGGCUGCAGAGGCCCAGUGCUCAGGCUCAUGCUCUGAUUGCGGUGAAGAUGAGCUUGGGCGAGCAGCAUAAUUA ...........(((((....)))))...((((((....))))))....(((((....((((((.(((((...((((((.......))).)))...)))))))))))..)))))....... (-33.74 = -34.78 + 1.04)

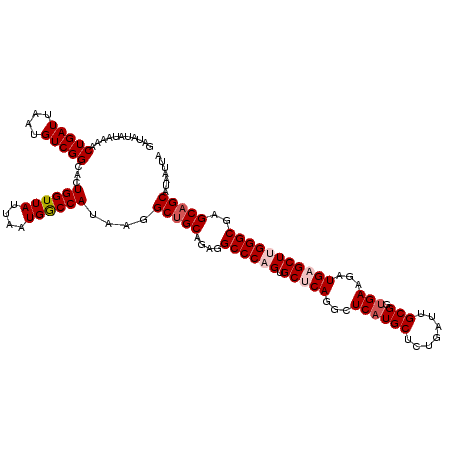
| Location | 1,561,624 – 1,561,744 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.75 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -28.26 |
| Energy contribution | -27.98 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1561624 120 - 20766785 UAAUUAUGCUGCUCGCCCAAGCUCAUCUUCACCGCAAUCAGAGCAUGAGCCUGUGCUCUGGGCCUCUGCAGCCUUAUGGCCAUUAAUAACCAGUGCCGACAUUAAUCAGUUUUAUAUAUC .......(((((..((((((((.((..((((..((.......)).))))..)).))).)))))....)))))....((((.(((.......)))))))...................... ( -29.40) >DroSec_CAF1 11365 120 - 1 UAAUUAUGCUGCUCGCCCGAGCUCAUCUUCACCGCAAUCAGAGCAUGAGAAUGAGCACUGGGCCUCUGCAGCCUUAUGGCCAUUAAUAACCAGUGCCGACAUUAAUCAGUUUUAUAUAUC .......(((((..(((((.((((((..(((..((.......)).)))..))))))..)))))....)))))....((((.(((.......)))))))...................... ( -32.50) >DroSim_CAF1 12898 120 - 1 UAAUUAUGCUGCUCGCCCGAGCUCAUCUUCACCGCAAUCAGAGCAUGAGCCUGAGCACUGGGCCUCUGCAGCCUUAUGGCCAUUAAUAACCAGUGCCGACAUUAAUCAGUUUUAUAUAUC .......(((((..(((((.(((((..((((..((.......)).))))..)))))..)))))....)))))....((((.(((.......)))))))...................... ( -30.70) >DroEre_CAF1 11740 120 - 1 UAAUUAUGCUGCUCGCCCAAGCUCAUCUUCGGCGCAAUCAGAGCAUGAGCCUGGGCACUGGGCCUCCGCAGCCUUAUGGCCAUUAAUAACCAGCGCCGACAUUAAUCAGUUUUAUAUAUC .....((((.((..(((((.((((((((..((.....))..)).)))))).))))).((((((....)).(((....))).........)))).)).).))).................. ( -33.80) >DroYak_CAF1 17728 120 - 1 UAAUUAUGCUGCUCGCCCAAGCUCAUCUUCACCGCAAUCAGAGCAUGAGCCUGAGCACUGGGCCUCUGCUGCCUUAUGGUCAUUAAUAACCAGCGCCGACAUUAAUCAGUUUUAUAUAUC .......((((...(((((.(((((..((((..((.......)).))))..)))))..))))).((.((.((....((((........)))))))).)).......)))).......... ( -27.60) >consensus UAAUUAUGCUGCUCGCCCAAGCUCAUCUUCACCGCAAUCAGAGCAUGAGCCUGAGCACUGGGCCUCUGCAGCCUUAUGGCCAUUAAUAACCAGUGCCGACAUUAAUCAGUUUUAUAUAUC .......(((((..(((((.(((((..((((..((.......)).))))..)))))..)))))....)))))....(((((...........).))))...................... (-28.26 = -27.98 + -0.28)

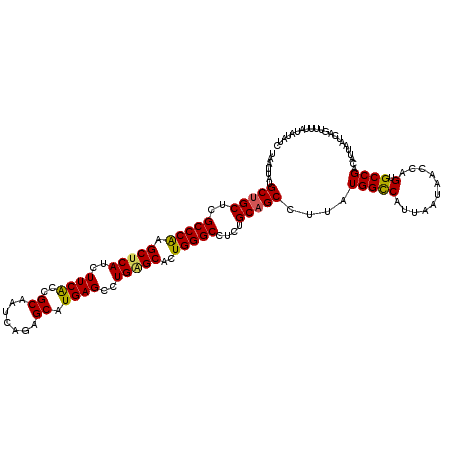
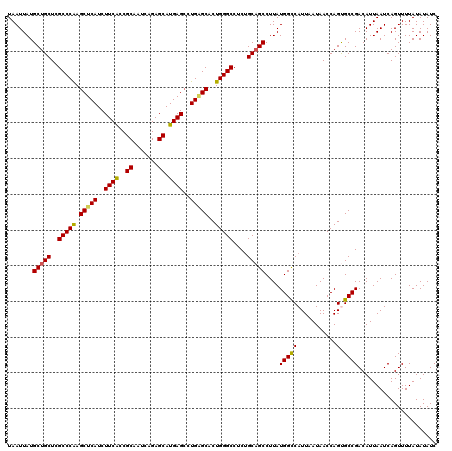
| Location | 1,561,664 – 1,561,768 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -37.56 |
| Consensus MFE | -29.14 |
| Energy contribution | -29.06 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
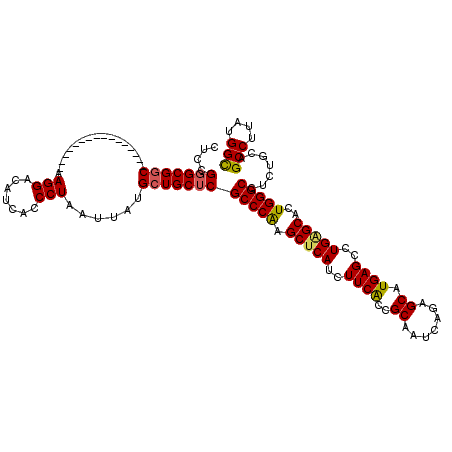
>2R_DroMel_CAF1 1561664 104 - 20766785 CUCCGGGCGGC----------------AAGGACAUCACCCUAAUUAUGCUGCUCGCCCAAGCUCAUCUUCACCGCAAUCAGAGCAUGAGCCUGUGCUCUGGGCCUCUGCAGCCUUAUGGC ....(((((((----------------((((.......))).....))))))))((((((((.((..((((..((.......)).))))..)).))).))))).......(((....))) ( -34.90) >DroSec_CAF1 11405 104 - 1 CUCCGGGCGAC----------------AAGGACUUCACCCUAAUUAUGCUGCUCGCCCGAGCUCAUCUUCACCGCAAUCAGAGCAUGAGAAUGAGCACUGGGCCUCUGCAGCCUUAUGGC ..(((((.((.----------------.......)).))).......(((((..(((((.((((((..(((..((.......)).)))..))))))..)))))....))))).....)). ( -35.50) >DroSim_CAF1 12938 104 - 1 CUCCGGGCGGC----------------AAGGACAUCACCCUAAUUAUGCUGCUCGCCCGAGCUCAUCUUCACCGCAAUCAGAGCAUGAGCCUGAGCACUGGGCCUCUGCAGCCUUAUGGC ....(((((((----------------((((.......))).....))))))))(((((.(((((..((((..((.......)).))))..)))))..))))).......(((....))) ( -36.20) >DroEre_CAF1 11780 120 - 1 CUCCGGGCGGCAAGGAGGCAAGGAGGCAAGGACAUCACCCUAAUUAUGCUGCUCGCCCAAGCUCAUCUUCGGCGCAAUCAGAGCAUGAGCCUGGGCACUGGGCCUCCGCAGCCUUAUGGC ..(((...(((..((((((..((.(((((((.......))).....))))....(((((.((((((((..((.....))..)).)))))).))))).))..))))))...)))...))). ( -48.00) >DroYak_CAF1 17768 104 - 1 CUCUGUGCGGC----------------AAGGACAUCAUCCUAAUUAUGCUGCUCGCCCAAGCUCAUCUUCACCGCAAUCAGAGCAUGAGCCUGAGCACUGGGCCUCUGCUGCCUUAUGGU ....(.(((((----------------(((((.....)))).....)))))).)(((((.(((((..((((..((.......)).))))..)))))..))))).......(((....))) ( -33.20) >consensus CUCCGGGCGGC________________AAGGACAUCACCCUAAUUAUGCUGCUCGCCCAAGCUCAUCUUCACCGCAAUCAGAGCAUGAGCCUGAGCACUGGGCCUCUGCAGCCUUAUGGC ....(((((((.................(((.......)))......)))))))(((((.(((((..((((..((.......)).))))..)))))..))))).......(((....))) (-29.14 = -29.06 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:57 2006