
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,260,626 – 8,260,763 |
| Length | 137 |
| Max. P | 0.974458 |

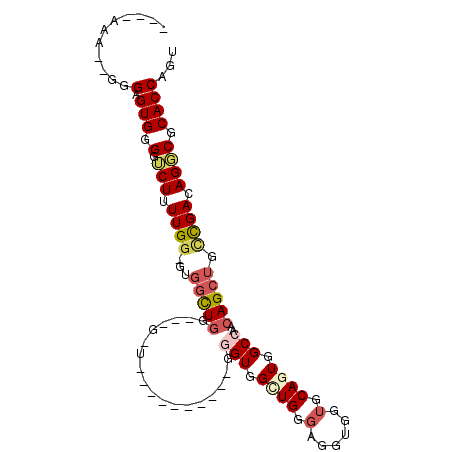
| Location | 8,260,626 – 8,260,723 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.91 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -14.50 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8260626 97 + 20766785 ----AAA--GGGAGUGGGGUCUUUUGG-GUGGUUGGGUGGCUGGUGGCUGUGGGGUGGCUGGGAGGUGGUGCAGUGGCCAACAGCUGCCGACAGGCGCACCAGU ----.((--((((......))))))..-.......(((((((((..(((((..(((.((((.(......).)))).))).)))))..))....))).))))... ( -37.10) >DroVir_CAF1 67546 88 + 1 GCUUUGGGGCUGUGUGGGGGCUUUUGUUGUUGUU-----GUUGUUU--------GUUG-UGGGUGCAG--GCAGUAGCCAACAGCUGUUGACAGCCGCACCAGU (((.((.((((((.(((.((((...((((..(((-----(((((((--------((..-.....))))--)))))))))))))))).))))))))).))..))) ( -30.30) >DroSec_CAF1 47862 82 + 1 ----AAA--GGGAGUGGGGACUUUUGG-GUGGCUGG---------------GGGGUGGCUGGGAGGUGGUGCAGUGGCCAACAGCUGCCGACAGGCGCACCAGU ----...--((((((....)))))).(-((((((.(---------------..((..((((.....((((......)))).))))..))..).))).))))... ( -31.90) >DroSim_CAF1 50205 79 + 1 ----AAA--GGGAGUGGGAUCUUUUGG-GUGGCUGG------------------GUGGUUGGGAGGUGGUGCAGUGGCCAACAGCUGCCGACAGGCGCACCAGU ----.((--((((......))))))((-(((.((((------------------(..((((.....((((......)))).))))..))..))).))).))... ( -27.80) >DroEre_CAF1 47837 87 + 1 ----AAU--GGGAGUGGGCUCUGUUGG-GUGGCUGGGUGGUU----------GGGUGGCUGGGAGGAGGUGCAGUGGCCAACAGCUGUCGACAGGCGCACCAGU ----...--..(.(((.((.((((((.-(..((((..(((((----------(.(((.((......)).)))..)))))).))))..))))))))).))))... ( -33.50) >DroYak_CAF1 49805 86 + 1 ----AAG--GGGAGUGGGGUCUGUUGG-GUGACUGGGUGGCU----------GGGUGG-UGGGAGGUGGUGCAGUGGCCAACAGCUGUCGACAGGCGCACCAGU ----...--((.....(.(((((((((-....)).((..(((----------(..(((-(.(..(......)..).)))).))))..))))))))).).))... ( -30.50) >consensus ____AAA__GGGAGUGGGGUCUUUUGG_GUGGCUGG___G_U__________GGGUGGCUGGGAGGUGGUGCAGUGGCCAACAGCUGCCGACAGGCGCACCAGU ...........(.(((.(.(((.((((...(((((..................(((.((((.(......).)))).)))..))))).)))).)))).))))... (-14.50 = -15.33 + 0.84)


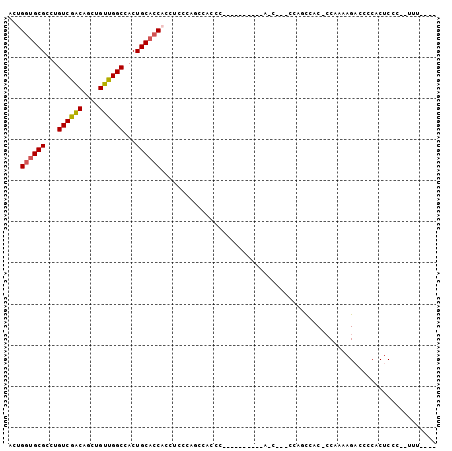
| Location | 8,260,626 – 8,260,723 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.91 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -15.04 |
| Energy contribution | -14.98 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8260626 97 - 20766785 ACUGGUGCGCCUGUCGGCAGCUGUUGGCCACUGCACCACCUCCCAGCCACCCCACAGCCACCAGCCACCCAACCAC-CCAAAAGACCCCACUCCC--UUU---- .((((((.(((....))).(((((((((.................))))....)))))))))))............-..................--...---- ( -22.23) >DroVir_CAF1 67546 88 - 1 ACUGGUGCGGCUGUCAACAGCUGUUGGCUACUGC--CUGCACCCA-CAAC--------AAACAAC-----AACAACAACAAAAGCCCCCACACAGCCCCAAAGC ...((((((((.((((((....))))))....))--).)))))..-....--------.......-----.................................. ( -20.60) >DroSec_CAF1 47862 82 - 1 ACUGGUGCGCCUGUCGGCAGCUGUUGGCCACUGCACCACCUCCCAGCCACCCC---------------CCAGCCAC-CCAAAAGUCCCCACUCCC--UUU---- ..(((((((...((((((....))))))...)))))))...............---------------........-..................--...---- ( -16.50) >DroSim_CAF1 50205 79 - 1 ACUGGUGCGCCUGUCGGCAGCUGUUGGCCACUGCACCACCUCCCAACCAC------------------CCAGCCAC-CCAAAAGAUCCCACUCCC--UUU---- ..(((((((...((((((....))))))...)))))))............------------------........-..................--...---- ( -16.50) >DroEre_CAF1 47837 87 - 1 ACUGGUGCGCCUGUCGACAGCUGUUGGCCACUGCACCUCCUCCCAGCCACCC----------AACCACCCAGCCAC-CCAACAGAGCCCACUCCC--AUU---- ...((((((...((((((....))))))...))))))...............----------..............-......(((....)))..--...---- ( -17.00) >DroYak_CAF1 49805 86 - 1 ACUGGUGCGCCUGUCGACAGCUGUUGGCCACUGCACCACCUCCCA-CCACCC----------AGCCACCCAGUCAC-CCAACAGACCCCACUCCC--CUU---- ..(((((((...((((((....))))))...))))))).......-......----------.........(((..-......))).........--...---- ( -18.50) >consensus ACUGGUGCGCCUGUCGACAGCUGUUGGCCACUGCACCACCUCCCAGCCACCC__________A_C___CCAGCCAC_CCAAAAGACCCCACUCCC__UUU____ ...((((((...((((((....))))))...))))))................................................................... (-15.04 = -14.98 + -0.05)


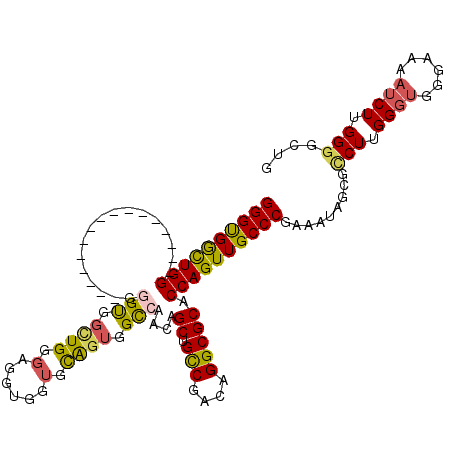
| Location | 8,260,645 – 8,260,763 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -47.35 |
| Consensus MFE | -32.83 |
| Energy contribution | -32.67 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8260645 118 + 20766785 GGGUGGUUGGGUGGCUGGUGGCUGUGGGGUGGCUGGGAGGUGGUGCAGUGGCCAACAGCUGCCGACAGGCGCACCAGUUGCCCGAAAUAGCGCCUUGGGUGGGAAAAUCUUGGGGCUG ..((..(((((..((((((((((((..(((.((((.(......).)))).))).))))).(((....))).)))))))..)))))....))((((..((.........))..)))).. ( -60.40) >DroSec_CAF1 47881 103 + 1 GGGUGGCUGG---------------GGGGUGGCUGGGAGGUGGUGCAGUGGCCAACAGCUGCCGACAGGCGCACCAGUUGCCCGAAAUAGCGACUUGGGUGGGAAAAUCUUGGGGCUG (((..(((((---------------...(((.(((...((..((...((.....)).))..))..))).))).)))))..)))....((((..((.((((......)))).)).)))) ( -44.60) >DroSim_CAF1 50224 100 + 1 GGGUGGCUGG------------------GUGGUUGGGAGGUGGUGCAGUGGCCAACAGCUGCCGACAGGCGCACCAGUUGCCCGAAAUAGCGCCUUGGGUGGGAAAAUCUUGGGGCUG (((..(((((------------------(((.(((...((..((...((.....)).))..))..))).))).)))))..)))....((((.(((.((((......)))).))))))) ( -45.60) >DroEre_CAF1 47856 105 + 1 GGGUGGCUGGGUGGUU----------GGGUGGCUGGGAGGAGGUGCAGUGGCCAACAGCUGUCGACAGGCGCACCAGUUGCCCCAAAUAGCGCCUUGGGUGGGAAAAUCUUGGGC--- (((..(((((((((((----------((.(.((((.(......).)))).))))))..(((....))).))).)))))..)))........((((.((((......)))).))))--- ( -43.90) >DroYak_CAF1 49824 104 + 1 GGGUGACUGGGUGGCU----------GGGUGG-UGGGAGGUGGUGCAGUGGCCAACAGCUGUCGACAGGCGCACCAGUUGCCCGAAAUAGCGCCUUGGGUGGGAAAAUCUUGGGC--- (((..((((((((.((----------(..(((-..(....((((......))))....)..))).))).))).)))))..)))........((((.((((......)))).))))--- ( -45.70) >DroAna_CAF1 50061 91 + 1 GGGCGACUGG------------------GCGACUGGGC---GGAGUGGGGGUCAACAGCUGCCGACAGGCGCACCAGUUGCCCGAAAUAGCUUCUUGGGUGG------CUUGGGUGGC ((((((((((------------------(((.((((((---((..((........)).)))))..))).))).))))))))))......(((.((..((...------))..)).))) ( -43.90) >consensus GGGUGGCUGG________________GGGUGGCUGGGAGGUGGUGCAGUGGCCAACAGCUGCCGACAGGCGCACCAGUUGCCCGAAAUAGCGCCUUGGGUGGGAAAAUCUUGGGGCUG ((((((((((.................(((.((((.(......).)))).)))....((.(((....))))).)))))))))).........(((.((((......)))).))).... (-32.83 = -32.67 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:33 2006