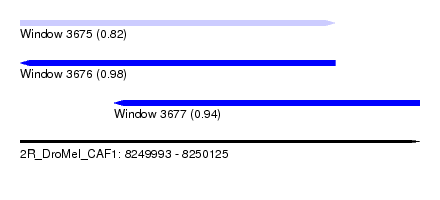
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,249,993 – 8,250,125 |
| Length | 132 |
| Max. P | 0.978459 |

| Location | 8,249,993 – 8,250,097 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.03 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8249993 104 + 20766785 UCA-UUUGAUUUUCCACUUUUUGGCCUGUGUCUGUG--UGUGUGUUUGCCCACUCGAAUGCCUCCGAGUGGCAAUUUUAUCGAAUUAUUUGCACUUAGC-UGCAAAUU ..(-((((((.((((((((...(((...((...(((--.((......)).))).))...)))...)))))).))....))))))).(((((((......-))))))). ( -25.20) >DroPse_CAF1 36868 84 + 1 -GCUGGUGC---------UUUUGGCUG--------------GCGUUUGCCCACUCGAAUGCCUCCAAGUGGCAAUUUUAUCGAAUUAUUUGCACUUAGCUUGCAAAUU -((((((((---------.(((((..(--------------(((((((......)))))))).)))))....(((((....)))))....)))).))))......... ( -23.40) >DroGri_CAF1 59068 81 + 1 --------G---------CUGUGGCUUGU----------GUGCUGUUGCCCACUCGAAUGCCUUCAAGUGCCAAUUUUAUCAAAUUAUUUGCACUUAACUCGCAAAUU --------(---------(...(((..((----------(.((....)).)))......)))...((((((.(((...........))).)))))).....))..... ( -16.90) >DroSim_CAF1 39708 99 + 1 UCA-UUCGAUU-------UUUUGGCCUGUGUGUGUGUGUGUUUGUUUGCCCACUCGAAUGCCUCCGAGUGGCAAUUUUAUCGAAUUAUUUGCACUUAGC-UGCAAAUU ...-.......-------.....((..((..(((((...(((((.(((((.(((((........))))))))))......)))))....)))))...))-.))..... ( -22.90) >DroEre_CAF1 37114 83 + 1 UGC-UUUGU---------UUUUGGCCU--------------GUGUUUGCCCACUCGAAUGCCUCCGAGUGGCAAUUUUAUCGAAUUAUUUGCACUUAGC-UGCAAAUU .((-(....---------....)))..--------------....(((((.(((((........))))))))))............(((((((......-))))))). ( -19.90) >DroYak_CAF1 38770 83 + 1 UGU-UUUGG---------UUUUGGCCU--------------GUGUUUGCCCACUCGAAUGCCUCCGAGUGGCAAUUUUAUCGAAUUAUUUGCACUUAGC-UGCAAAUU ...-...((---------(....))).--------------....(((((.(((((........))))))))))............(((((((......-))))))). ( -20.40) >consensus UGA_UUUGA_________UUUUGGCCU______________GUGUUUGCCCACUCGAAUGCCUCCGAGUGGCAAUUUUAUCGAAUUAUUUGCACUUAGC_UGCAAAUU .............................................(((((.(((((........))))))))))............(((((((.......))))))). (-14.10 = -14.77 + 0.67)

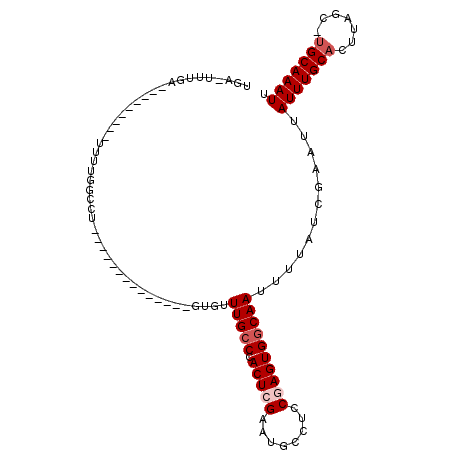
| Location | 8,249,993 – 8,250,097 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.03 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -19.63 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8249993 104 - 20766785 AAUUUGCA-GCUAAGUGCAAAUAAUUCGAUAAAAUUGCCACUCGGAGGCAUUCGAGUGGGCAAACACACA--CACAGACACAGGCCAAAAAGUGGAAAAUCAAA-UGA .(((((((-......))))))).....(((....((((((((((((....))))))).))))).......--............(((.....)))...)))...-... ( -24.60) >DroPse_CAF1 36868 84 - 1 AAUUUGCAAGCUAAGUGCAAAUAAUUCGAUAAAAUUGCCACUUGGAGGCAUUCGAGUGGGCAAACGC--------------CAGCCAAAA---------GCACCAGC- .........(((..((((.....((((((......((((.......)))).)))))).(((....))--------------)........---------)))).)))- ( -21.30) >DroGri_CAF1 59068 81 - 1 AAUUUGCGAGUUAAGUGCAAAUAAUUUGAUAAAAUUGGCACUUGAAGGCAUUCGAGUGGGCAACAGCAC----------ACAAGCCACAG---------C-------- ...(((.(.(((..((((...((((((....)))))).((((((((....))))))))(....).))))----------...)))).)))---------.-------- ( -19.10) >DroSim_CAF1 39708 99 - 1 AAUUUGCA-GCUAAGUGCAAAUAAUUCGAUAAAAUUGCCACUCGGAGGCAUUCGAGUGGGCAAACAAACACACACACACACAGGCCAAAA-------AAUCGAA-UGA .(((((((-......))))))).(((((((....((((((((((((....))))))).))))).............(.....).......-------.))))))-).. ( -26.20) >DroEre_CAF1 37114 83 - 1 AAUUUGCA-GCUAAGUGCAAAUAAUUCGAUAAAAUUGCCACUCGGAGGCAUUCGAGUGGGCAAACAC--------------AGGCCAAAA---------ACAAA-GCA .(((((((-......))))))).............(((((((((((....))))))))(((......--------------..)))....---------.....-))) ( -22.70) >DroYak_CAF1 38770 83 - 1 AAUUUGCA-GCUAAGUGCAAAUAAUUCGAUAAAAUUGCCACUCGGAGGCAUUCGAGUGGGCAAACAC--------------AGGCCAAAA---------CCAAA-ACA .(((((((-......)))))))............((((((((((((....))))))).)))))....--------------.((......---------))...-... ( -22.70) >consensus AAUUUGCA_GCUAAGUGCAAAUAAUUCGAUAAAAUUGCCACUCGGAGGCAUUCGAGUGGGCAAACAC______________AGGCCAAAA_________CCAAA_GCA .(((((((.......)))))))............((((((((((((....))))))).)))))............................................. (-19.63 = -19.30 + -0.33)
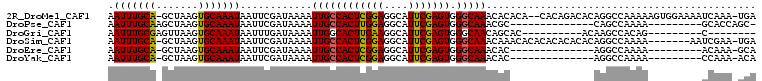
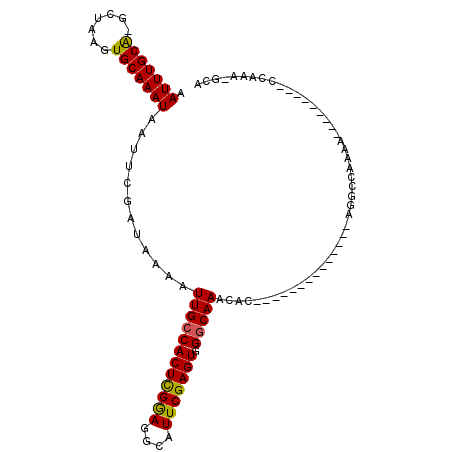
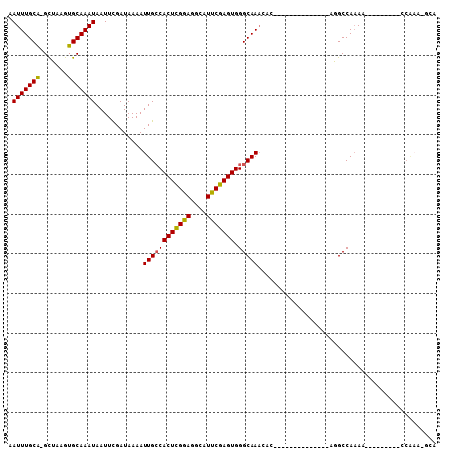
| Location | 8,250,024 – 8,250,125 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.03 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -14.50 |
| Energy contribution | -14.12 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
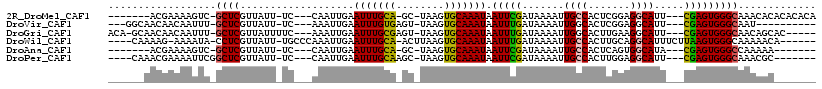
>2R_DroMel_CAF1 8250024 101 - 20766785 -------ACGAAAAGUC-GCUCGUUAUU-UC---CAAUUGAAUUUGCA-GC-UAAGUGCAAAUAAUUCGAUAAAAUUGCCACUCGGAGGCAUU---CGAGUGGGCAAACACACACACA -------((((......-..))))....-..---..((((((((((((-..-....))))...))))))))....((((((((((((....))---))))).)))))........... ( -25.30) >DroVir_CAF1 52719 96 - 1 ---GGCAACAACAAUUU-GCUCGUUAUU-UC---AAAUUGAAUUUGUGAGU-UAAGUGCAAAUAAUUUGAUAAAAUUGGCACUCGGAGGCAUU---CGAGUGGGCAAU---------- ---(....)......((-((((((((..-((---(((((..((((((....-.....)))))))))))))......))))(((((((....))---))))))))))).---------- ( -24.30) >DroGri_CAF1 59080 104 - 1 ACA-GCAACAACAAUUU-GCUCGUUAUUUUC---AAAUUGAAUUUGCGAGU-UAAGUGCAAAUAAUUUGAUAAAAUUGGCACUUGAAGGCAUU---CGAGUGGGCAACAGCAC----- ...-.............-((((((....(((---.....)))...))))))-...((((...((((((....)))))).((((((((....))---))))))(....).))))----- ( -24.90) >DroWil_CAF1 50335 104 - 1 ----CAAAAG-AAAAUA-CCUCGUUAUU-UGCCCAAAUUGAAUUUGCA-ACUUAAGUGCAAAUAAUUUGAUAAAAUUGCCACUUGCAGGCAUUUCUUAAGUGGGCAAAAACA------ ----......-......-....(((.((-((((((....(((..(((.-...((((((....((((((....)))))).))))))...))).))).....))))))))))).------ ( -25.10) >DroAna_CAF1 39808 94 - 1 -------ACGAAAAGUC-GCUCGUUAUU-UC---CAAUUGAAUUUGCA-GC-UAAGUGCAAAUAAUUCGAUAAAAUUGCCACUCAGUGGCAUA---CGAGUGGGCCAAAAA------- -------........((-((((((....-..---..((((((((((((-..-....))))...)))))))).....((((((...)))))).)---)))))))........------- ( -25.70) >DroPer_CAF1 36616 99 - 1 ----CAAACGAAAAUUCGGCUCGUUAUU-UC---CAAUUGAAUUUGCAAGC-UAAGUGCAAAUAAUUCGAUAAAAUUGCCACUUGGAGGCAUU---CGAGUGGGCAAACGC------- ----..(((((.........)))))...-..---..((((((((((((...-....))))...))))))))....((((((((((((....))---))))).)))))....------- ( -23.80) >consensus ______AACGAAAAUUC_GCUCGUUAUU_UC___AAAUUGAAUUUGCA_GC_UAAGUGCAAAUAAUUCGAUAAAAUUGCCACUCGGAGGCAUU___CGAGUGGGCAAAAAC_______ ..................((((...................(((((((........))))))).(((((.......((((.......)))).....)))))))))............. (-14.50 = -14.12 + -0.39)
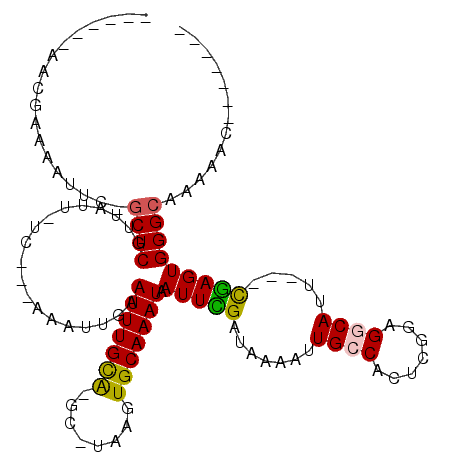
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:28 2006