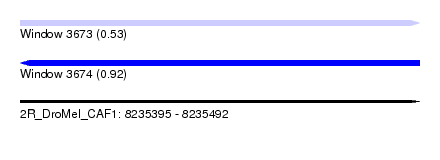
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,235,395 – 8,235,492 |
| Length | 97 |
| Max. P | 0.921367 |

| Location | 8,235,395 – 8,235,492 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -20.54 |
| Consensus MFE | -10.18 |
| Energy contribution | -10.40 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
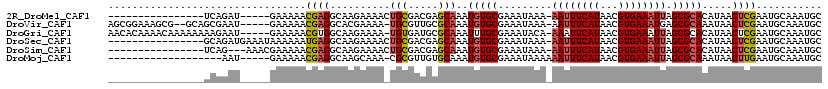
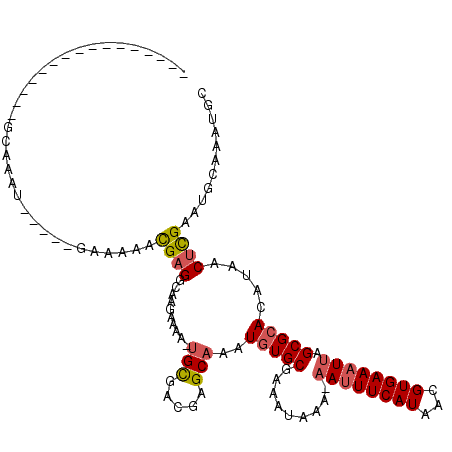
>2R_DroMel_CAF1 8235395 97 + 20766785 GCAUUUGCAUUCGAGUUAUGUGCGCUAAUUUCACGUUAUGAAAUU-UUUAUUUCGCACAUUUGCUCGUCGCAGUUUUCUUGCCUCGUUUUUC-----AUCUGA---------------- (((.((((...(((((.(((((((..(((((((.....)))))))-.......)))))))..)))))..))))......)))..........-----......---------------- ( -24.00) >DroVir_CAF1 34317 110 + 1 GCAUUUGCAUUCGAGUUAUUUGCGCUCAUUUCACGUUAUGAAAUU-UUUAUUUCGCACAUUUGCGCAACGCA-UUUUCGUGCCUCGUUUUUC-----AUUCGCUGC--CGCUUUCCGCU ......((...((((....((((((........((..((((....-.))))..)).......)))))).(((-(....))))))))......-----....((...--.)).....)). ( -18.96) >DroGri_CAF1 39866 112 + 1 GCAUUUGCAUUCGAGUUAUGUGCGCUAAUUUCACGUUAUGAAUUU-UGUAUUUCGCAAAUUUGCGCAUCACA-UUUUCUUGCCACGUUUUUC-----AUUCUUUUUUUUGUUUUGUGUU ((....))....(((..(((((.((((((.....)))).(((...-(((..(.((((....)))).)..)))-..)))..)))))))..)))-----...................... ( -16.70) >DroSec_CAF1 22801 102 + 1 GCAUUUGCAUUCGAGUUAUGUGCGCUAAUUUCACGUUAUGAAAUU-UUUAUUUCGCACAUUUGCUCGUCGCAGUUUUCUUGCCUCAUUUUUUAUUUCAUCUGC---------------- (((.((((...(((((.(((((((..(((((((.....)))))))-.......)))))))..)))))..))))......))).....................---------------- ( -24.00) >DroSim_CAF1 24823 99 + 1 GCAUUUGCAUUCGAGUUAUGUGCGCUAAUUUCACGUUAUGAAAUU-UUUAUUUCGCACAUUUGCUCGUCGCAGUUUUCUUGCCUCGUUUUUCGUUU---CUGA---------------- (((.((((...(((((.(((((((..(((((((.....)))))))-.......)))))))..)))))..))))......)))..............---....---------------- ( -24.00) >DroMoj_CAF1 39533 94 + 1 GCAUUUGCAUUCAAGUUAUUUGCGCUAAUUUCACGUUAUGAAAUUUUUUAUUUCGCACAUUUGCACAACGCG-UUUGCUUGCCUCGUUUUUC-----AUU------------------- ((....))...(((((.....((((.(((((((.....))))))).........(((....))).....)))-)..)))))...........-----...------------------- ( -15.60) >consensus GCAUUUGCAUUCGAGUUAUGUGCGCUAAUUUCACGUUAUGAAAUU_UUUAUUUCGCACAUUUGCUCAUCGCA_UUUUCUUGCCUCGUUUUUC_____AUCUGA________________ ((....))...((((.....((((...............(((((.....)))))(((....)))....)))).....))))...................................... (-10.18 = -10.40 + 0.22)

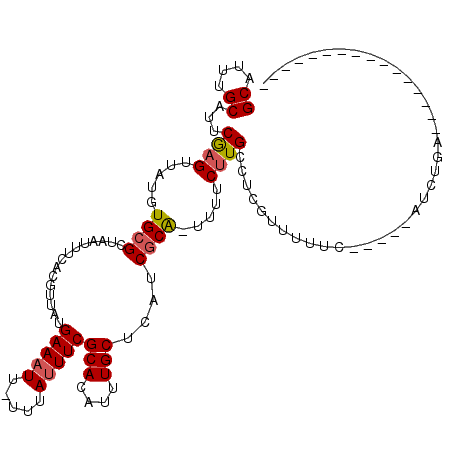
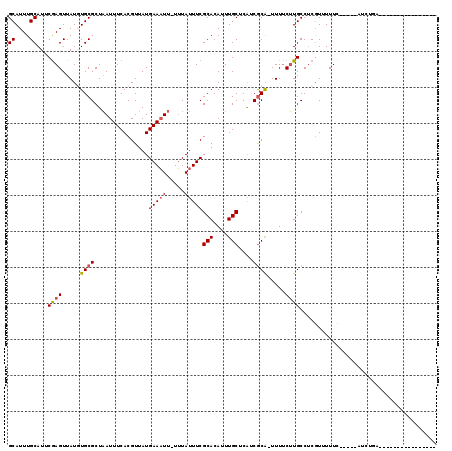
| Location | 8,235,395 – 8,235,492 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -12.66 |
| Energy contribution | -13.08 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8235395 97 - 20766785 ----------------UCAGAU-----GAAAAACGAGGCAAGAAAACUGCGACGAGCAAAUGUGCGAAAUAAA-AAUUUCAUAACGUGAAAUUAGCGCACAUAACUCGAAUGCAAAUGC ----------------......-----......((((..........(((.....))).(((((((.......-((((((((...))))))))..)))))))..))))........... ( -21.00) >DroVir_CAF1 34317 110 - 1 AGCGGAAAGCG--GCAGCGAAU-----GAAAAACGAGGCACGAAAA-UGCGUUGCGCAAAUGUGCGAAAUAAA-AAUUUCAUAACGUGAAAUGAGCGCAAAUAACUCGAAUGCAAAUGC .(((....((.--...))....-----......(((((((......-))).((((((...(((.....)))..-.(((((((...)))))))..))))))....))))..)))...... ( -24.10) >DroGri_CAF1 39866 112 - 1 AACACAAAACAAAAAAAAGAAU-----GAAAAACGUGGCAAGAAAA-UGUGAUGCGCAAAUUUGCGAAAUACA-AAAUUCAUAACGUGAAAUUAGCGCACAUAACUCGAAUGCAAAUGC ..(((.................-----.......)))(((.((...-((((.(((((.((((((((.......-..........))).))))).)))))))))..))...)))...... ( -16.79) >DroSec_CAF1 22801 102 - 1 ----------------GCAGAUGAAAUAAAAAAUGAGGCAAGAAAACUGCGACGAGCAAAUGUGCGAAAUAAA-AAUUUCAUAACGUGAAAUUAGCGCACAUAACUCGAAUGCAAAUGC ----------------(((..................(((.......)))..((((...(((((((.......-((((((((...))))))))..)))))))..))))..)))...... ( -22.80) >DroSim_CAF1 24823 99 - 1 ----------------UCAG---AAACGAAAAACGAGGCAAGAAAACUGCGACGAGCAAAUGUGCGAAAUAAA-AAUUUCAUAACGUGAAAUUAGCGCACAUAACUCGAAUGCAAAUGC ----------------....---..........((((..........(((.....))).(((((((.......-((((((((...))))))))..)))))))..))))........... ( -21.00) >DroMoj_CAF1 39533 94 - 1 -------------------AAU-----GAAAAACGAGGCAAGCAAA-CGCGUUGUGCAAAUGUGCGAAAUAAAAAAUUUCAUAACGUGAAAUUAGCGCAAAUAACUUGAAUGCAAAUGC -------------------...-----..........(((..(((.-....((((((...(((.....)))...((((((((...)))))))).)))))).....)))..)))...... ( -17.10) >consensus ________________GCAAAU_____GAAAAACGAGGCAAGAAAA_UGCGACGAGCAAAUGUGCGAAAUAAA_AAUUUCAUAACGUGAAAUUAGCGCACAUAACUCGAAUGCAAAUGC .................................((((..........(((.....)))..(((((.........((((((((...)))))))).))))).....))))........... (-12.66 = -13.08 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:26 2006