| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,234,078 – 8,234,216 |
| Length | 138 |
| Max. P | 0.999161 |

| Location | 8,234,078 – 8,234,191 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -21.76 |
| Energy contribution | -21.72 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8234078 113 + 20766785 AUUUGGCACGGCAUCUCAAACCAAGUAAUUAAACAUCAUUCC-GAAAGCAUUUCCAUCAGUAAUGCCAAUCUAAUCCAAAGUAGAUUGGCAAAUACAGCAUUUUAAUUGGAUGG------ ((((((..............))))))...............(-....).....(((((.(((.((((((((((........))))))))))..)))..((.......)))))))------ ( -22.84) >DroSec_CAF1 21509 114 + 1 AUUUGGCACGGCAUCUCAUCCCAAGUAAUUAAACCUCAUUCCCAAAAGCAUUUCCAUCAGUAAUGCCAAUCUAAUCCAAAGUAGAUUGGCAAAUACAGCAUUUUAAUUGGAUGG------ ((((((...((....))...))))))...........................(((((.(((.((((((((((........))))))))))..)))..((.......)))))))------ ( -22.40) >DroSim_CAF1 23536 114 + 1 AUUUGGCACGGCAUCUCAUCCCAAGUAAUUAAACCCCAUUCCCAAAAGCAUUUCCAUCAGUAAUGCCAAUCUAAUCCAAAGUAGAUUGGCAAAUACAGCAUUUUAAUUGGAUGG------ ((((((...((....))...))))))...........................(((((.(((.((((((((((........))))))))))..)))..((.......)))))))------ ( -22.40) >DroEre_CAF1 21091 113 + 1 AUUUGGCUCGGCAAGGCAUCCCAAGUAAUUAAACAUCAUUCC-GAAAGCAUUUCCCUCAGUAAUGCCAAUCUAAUCCAGAGUGGAUUGGCAAAUACAGCAUUUUAAUUGGAUGG------ ((((((....((...))...)))))).......((((....(-....)...........(((.((((((((((........))))))))))..))).............)))).------ ( -24.00) >DroYak_CAF1 22044 119 + 1 AUUUGGCUUGGCUACGCAUCCCAAGUAAUUAAACUUCAGUCC-GAAAGCAUUCCCAUCAGUAAUGCCAAUCUAAUCCAGAGUGGAUUGGCAAAUACAGCAUUUUAAUUGGAUGGCAAAUG (((((((((((.........))))))................-..........(((((.(((.((((((((((........))))))))))..)))..((.......)))))))))))). ( -28.00) >consensus AUUUGGCACGGCAUCUCAUCCCAAGUAAUUAAACAUCAUUCC_GAAAGCAUUUCCAUCAGUAAUGCCAAUCUAAUCCAAAGUAGAUUGGCAAAUACAGCAUUUUAAUUGGAUGG______ ((((((....(.....)...))))))...........................(((((.(((.((((((((((........))))))))))..)))..((.......)))))))...... (-21.76 = -21.72 + -0.04)

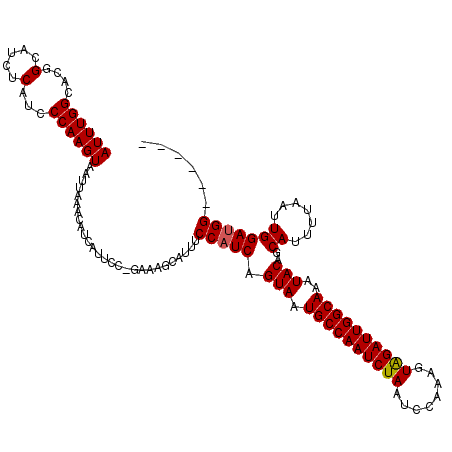
| Location | 8,234,118 – 8,234,216 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -24.36 |
| Consensus MFE | -21.56 |
| Energy contribution | -21.36 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8234118 98 + 20766785 CC-GAAAGCAUUUCCAUCAGUAAUGCCAAUCUAAUCCAAAGUAGAUUGGCAAAUACAGCAUUUUAAUUGGAUGG-------CAAAAAUGGGAUUUAGCAC----------AUUCAA ..-....((..((((((..(((.((((((((((........))))))))))..)))..(((((.....))))).-------.....))))))....))..----------...... ( -23.50) >DroSec_CAF1 21549 99 + 1 CCCAAAAGCAUUUCCAUCAGUAAUGCCAAUCUAAUCCAAAGUAGAUUGGCAAAUACAGCAUUUUAAUUGGAUGG-------CAAAAAUGGGAUUUAGCAC----------AUUCAA ((((.........(((((.(((.((((((((((........))))))))))..)))..((.......)))))))-------......)))).........----------...... ( -24.36) >DroSim_CAF1 23576 99 + 1 CCCAAAAGCAUUUCCAUCAGUAAUGCCAAUCUAAUCCAAAGUAGAUUGGCAAAUACAGCAUUUUAAUUGGAUGG-------CAAAAAUGGGUUUUAGCAC----------AUUCAA ((((.........(((((.(((.((((((((((........))))))))))..)))..((.......)))))))-------......)))).........----------...... ( -23.86) >DroEre_CAF1 21131 98 + 1 CC-GAAAGCAUUUCCCUCAGUAAUGCCAAUCUAAUCCAGAGUGGAUUGGCAAAUACAGCAUUUUAAUUGGAUGG-------CAAAAAUGGGUUUUAGCAC----------AUUCAA .(-....)...........(((.((((((((((........))))))))))..)))..........((((((((-------(.((((....)))).)).)----------)))))) ( -23.20) >DroYak_CAF1 22084 115 + 1 CC-GAAAGCAUUCCCAUCAGUAAUGCCAAUCUAAUCCAGAGUGGAUUGGCAAAUACAGCAUUUUAAUUGGAUGGCAAAUGGCAAAAAUGGGUUUUAGCACAUUCGAGCACAUUCAA ..-(((..((((.(((((.(((.((((((((((........))))))))))..)))..((.......)))))))..))))((...((((.((....)).))))...))...))).. ( -26.90) >consensus CC_GAAAGCAUUUCCAUCAGUAAUGCCAAUCUAAUCCAAAGUAGAUUGGCAAAUACAGCAUUUUAAUUGGAUGG_______CAAAAAUGGGUUUUAGCAC__________AUUCAA .......((...(((((..(((.((((((((((........))))))))))..)))..(((((.....))))).............))))).....)).................. (-21.56 = -21.36 + -0.20)

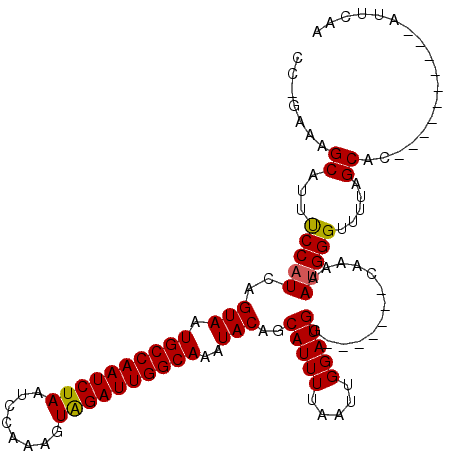
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:24 2006