| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,211,077 – 8,211,215 |
| Length | 138 |
| Max. P | 0.795408 |

| Location | 8,211,077 – 8,211,181 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -20.72 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8211077 104 - 20766785 UCAC--------CACUGUGCAUCGCAACACCUUGGCCAAUGCCAAUUAGUUGGCCUU--CUGGCUUCUG--CUGGCCUGGCCAAAUUUCAAUUAACGUCAAGAGCAAUCAACGAAA ....--------.........(((.......(((((((..((((..(((..((((..--..)))).)))--.)))).))))))).................((....))..))).. ( -26.90) >DroSec_CAF1 6538 104 - 1 UAAC--------CACUGAGCAUCGCAACACCUUGGCCAAUGCCAAUUAGUUGGCCUU--CUGGCUUCUG--CUGUCCUGGCCAAAUUUCAAUUAACGUCAAGAGCAAUCAACGAAA ....--------.........(((.......(((((((..(((((....)))))...--..(((....)--))....))))))).................((....))..))).. ( -22.10) >DroSim_CAF1 6461 104 - 1 UAAC--------CACUGUGCAUCGCAACACCUUGGCCAAUGCCAAUUAGUUGGCCUU--CUGGCUUUUG--CUGGCCUGGCCAAAUUUCAAUUAACGUCAAGAGCAAUCAACGAAA ....--------.....(((.((...((...(((((....)))))....((((((..--(..((....)--)..)...))))))............))...))))).......... ( -25.70) >DroEre_CAF1 6373 102 - 1 UAGC--------CAC--AGCAUCGCAACACCUUGGCCAAUGCCAAUUAGUUGGCCUU--CUGGCUUCUC--CGGGCCUGGCCAAAUUUCAAUUAACGUCAAGAGCAAUCAACGAAA ..((--------...--.)).(((.......(((((((..(((....((..((((..--..)))).)).--..))).))))))).................((....))..))).. ( -25.90) >DroYak_CAF1 6451 109 - 1 UAGCUAUCUCAGCAC--AGCAUCGCAACACCUUGGCCACUGCCAAUUAGUUGGCCUU--CUGGCUUCUC--CUGG-CUGGCCAAAUUUCAAUUAACGUCAAGAGCAAUCAACGAAA ..(((.....)))..--....(((.......(((((((..((((...((..((((..--..)))).)).--.)))-)))))))).................((....))..))).. ( -27.70) >DroAna_CAF1 8688 93 - 1 -----------------------CCAACACCUUGGUCACUGCCAAUUAGCCGGCUUCCUCCGGGUCCUGGCCUGGCCUGGCCAAAUUUCAAUUAACGUCAAGAGCAAUCAACGAAA -----------------------........(((((((..((((....(((((..(((...)))..))))).)))).)))))))...........(((...((....)).)))... ( -25.30) >consensus UAAC________CAC__AGCAUCGCAACACCUUGGCCAAUGCCAAUUAGUUGGCCUU__CUGGCUUCUG__CUGGCCUGGCCAAAUUUCAAUUAACGUCAAGAGCAAUCAACGAAA ...............................(((((((..((((..(((..((((......)))).)))...)))).)))))))...........(((...((....)).)))... (-20.72 = -21.30 + 0.58)


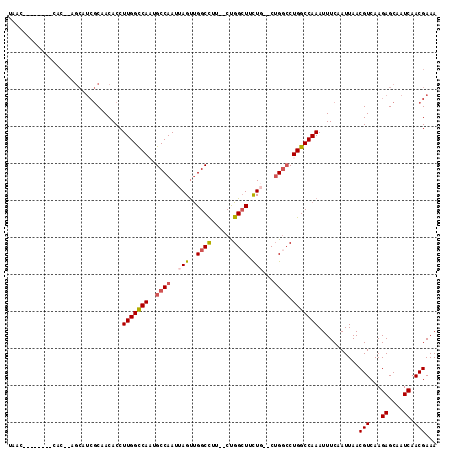
| Location | 8,211,117 – 8,211,215 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8211117 98 + 20766785 GCCAG--CAGAAGCCAG--AAGGCCAACUAAUUGGCAUUGGCCAAGGUGUUGCGAUGCACAGUG--------GUGAU-CUCUAUCUCCAUUACA--UUAGUUGGCCUACACUC ....(--(....))..(--.((((((((((((((((....))))).((((......))))((((--------(.(((-....))).)))))...--))))))))))).).... ( -34.20) >DroSec_CAF1 6578 96 + 1 GACAG--CAGAAGCCAG--AAGGCCAACUAAUUGGCAUUGGCCAAGGUGUUGCGAUGCUCAGUG--------GUUAU-CUCUAUCUCCAUUACA--UUAGUUGGCCUA--CUC ....(--(....)).((--.((((((((((((((((....)))))((.(..(.((((((....)--------).)))-).)...).))......--))))))))))).--)). ( -29.00) >DroSim_CAF1 6501 96 + 1 GCCAG--CAAAAGCCAG--AAGGCCAACUAAUUGGCAUUGGCCAAGGUGUUGCGAUGCACAGUG--------GUUAU-CUCUAUCUCCAUUACA--UUAGUUGGCCUA--CUC ....(--(....)).((--.(((((((((((((((...((((((..((((......))))..))--------)))).-..)))..........)--))))))))))).--)). ( -30.90) >DroEre_CAF1 6413 94 + 1 GCCCG--GAGAAGCCAG--AAGGCCAACUAAUUGGCAUUGGCCAAGGUGUUGCGAUGCU--GUG--------GCUAU-CUCUAUCUCCAUUACA--UUAGUUGGCCCA--CUC ....(--(.....))..--..(((((((((((((((....)))))((.(..(.(((((.--...--------)).))-).)...).))......--))))))))))..--... ( -29.40) >DroYak_CAF1 6491 102 + 1 -CCAG--GAGAAGCCAG--AAGGCCAACUAAUUGGCAGUGGCCAAGGUGUUGCGAUGCU--GUGCUGAGAUAGCUAUCCUCUAGCUCCAUUACA--UUAGUUGGCCUG--CUC -...(--(.....))((--.((((((((((((((((....)))))((.((((.((((((--((......))))).)))...)))).))......--))))))))))).--)). ( -36.50) >DroAna_CAF1 8728 85 + 1 GCCAGGCCAGGACCCGGAGGAAGCCGGCUAAUUGGCAGUGACCAAGGUGUUGG--------------------------CUUAUCUUCAUUACACAGUUGUUGGCCUU--CUC ...(((((((....)(((((((((((((...((((......))))...)))))--------------------------))).))))).............)))))).--... ( -30.30) >consensus GCCAG__CAGAAGCCAG__AAGGCCAACUAAUUGGCAUUGGCCAAGGUGUUGCGAUGCU__GUG________GUUAU_CUCUAUCUCCAUUACA__UUAGUUGGCCUA__CUC ....................((((((((((((((((....)))))((.(...................................).))........)))))))))))...... (-18.76 = -19.18 + 0.42)

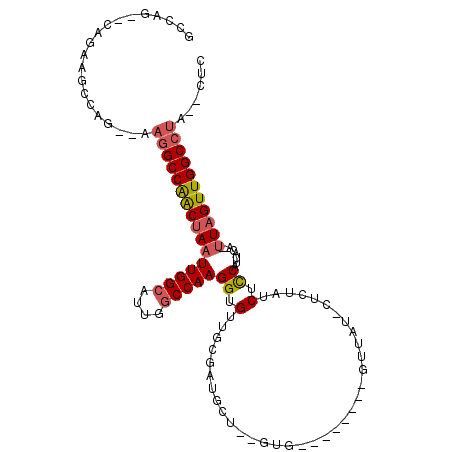
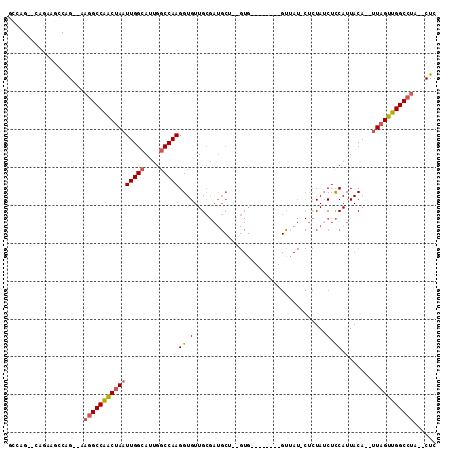
| Location | 8,211,117 – 8,211,215 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -29.69 |
| Consensus MFE | -17.99 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8211117 98 - 20766785 GAGUGUAGGCCAACUAA--UGUAAUGGAGAUAGAG-AUCAC--------CACUGUGCAUCGCAACACCUUGGCCAAUGCCAAUUAGUUGGCCUU--CUGGCUUCUG--CUGGC ......(((((((((((--((((.(((.(((....-))).)--------))...))))..........(((((....)))))))))))))))).--(..((....)--)..). ( -33.30) >DroSec_CAF1 6578 96 - 1 GAG--UAGGCCAACUAA--UGUAAUGGAGAUAGAG-AUAAC--------CACUGAGCAUCGCAACACCUUGGCCAAUGCCAAUUAGUUGGCCUU--CUGGCUUCUG--CUGUC .((--.(((((((((((--((..(((....(((.(-.....--------).)))..)))..)).....(((((....)))))))))))))))).--))(((....)--))... ( -28.70) >DroSim_CAF1 6501 96 - 1 GAG--UAGGCCAACUAA--UGUAAUGGAGAUAGAG-AUAAC--------CACUGUGCAUCGCAACACCUUGGCCAAUGCCAAUUAGUUGGCCUU--CUGGCUUUUG--CUGGC ...--.(((((((((((--((..(((...((((.(-.....--------).)))).)))..)).....(((((....)))))))))))))))).--(..((....)--)..). ( -30.80) >DroEre_CAF1 6413 94 - 1 GAG--UGGGCCAACUAA--UGUAAUGGAGAUAGAG-AUAGC--------CAC--AGCAUCGCAACACCUUGGCCAAUGCCAAUUAGUUGGCCUU--CUGGCUUCUC--CGGGC ...--.(((((((((((--(((..(((..((....-))..)--------)).--.)))..........(((((....))))))))))))))))(--((((.....)--)))). ( -30.00) >DroYak_CAF1 6491 102 - 1 GAG--CAGGCCAACUAA--UGUAAUGGAGCUAGAGGAUAGCUAUCUCAGCAC--AGCAUCGCAACACCUUGGCCACUGCCAAUUAGUUGGCCUU--CUGGCUUCUC--CUGG- (((--((((((((((((--(.....(((((((.....)))))......((..--......))....)).((((....)))))))))))))))).--...))))...--....- ( -33.10) >DroAna_CAF1 8728 85 - 1 GAG--AAGGCCAACAACUGUGUAAUGAAGAUAAG--------------------------CCAACACCUUGGUCACUGCCAAUUAGCCGGCUUCCUCCGGGUCCUGGCCUGGC (((--.(((((.......((((............--------------------------...)))).(((((....)))))......))))).)))(((((....))))).. ( -22.26) >consensus GAG__UAGGCCAACUAA__UGUAAUGGAGAUAGAG_AUAAC________CAC__AGCAUCGCAACACCUUGGCCAAUGCCAAUUAGUUGGCCUU__CUGGCUUCUG__CUGGC ......((((((((((...((..(((..............................)))..)).....(((((....))))).)))))))))).................... (-17.99 = -18.58 + 0.58)

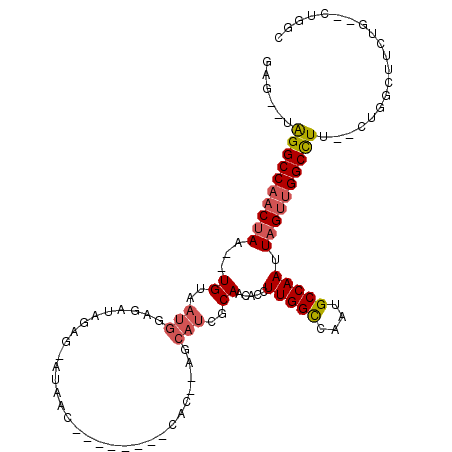

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:19 2006