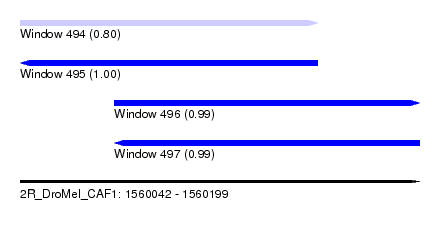
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,560,042 – 1,560,199 |
| Length | 157 |
| Max. P | 0.999836 |

| Location | 1,560,042 – 1,560,159 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.82 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -30.40 |
| Energy contribution | -29.76 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
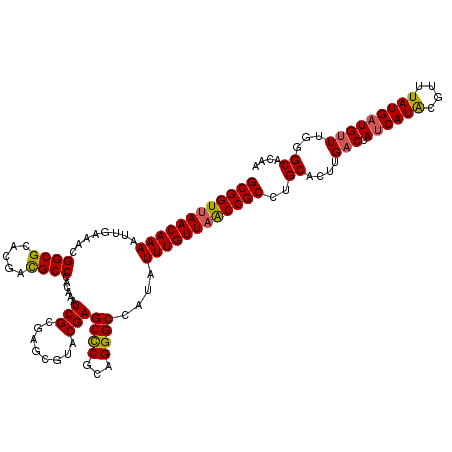
>2R_DroMel_CAF1 1560042 117 + 20766785 ---CUUUCCUAACAUCACCUUUCGGUGCCAUAAAAGUUUUGCGGUUAACAAAAUUGAAACGGCGCACGACGCCCACAAAUCGCUAGCGUACGAGCCCGCAGGGCCACAUUUGUUAACCGC ---............((((....)))).............((((((((((((........((((.....))))......(((........)))((((...))))....)))))))))))) ( -31.90) >DroSec_CAF1 9782 117 + 1 ---CUUUUCUAACAUCGCCUUUCGGCGCCAUAAAAGUUUUGCGGUUAACAAAAUUGAAACGGCGCACGACGCCCACAAAUCGCGAGCGUGCGAGCCCGCAGGGCCAUAUUUGUUAACCGC ---(((((.......((((....))))....)))))....((((((((((((..((......((((((.(((.........)))..)))))).((((...))))))..)))))))))))) ( -40.00) >DroSim_CAF1 11279 120 + 1 AUCCCUUCCUAACAUCGCCUUUCGGUGCCAUAAAAGUUUUGCGGUUAACAAAAUUGAAACGGCGCACGACGCCCACAAAUCGCGAGCGUACGAGCCCGCAGGGCCAUAUUUGUUAACCGC ...............((((....)))).............((((((((((((..((....((((.....))))........(((.((......)).))).....))..)))))))))))) ( -33.60) >DroEre_CAF1 10178 116 + 1 ---CCUUU-GGGCCUCACCUUUCGGUGCUAUAAAAGUUAUGCGGUGAACAAAAUGGAAACGGCGCACGACGCCCACAAAUCGCAAACCUACGAGCCCGCAGGGCCAUAUUUGUUAGCCGC ---.((((-.(((...(((....))))))...))))....(((((.((((((((((....((((.....)))).............(((.((....)).))).)))).)))))).))))) ( -33.30) >DroYak_CAF1 16342 117 + 1 ---CUUUCCGAGCCUUACCUUUAAGUGCCAUAAAAGUUUUGCGGUUAACAAAAUUACAAAGGCGCACGAUGCCCACAAAUCGCGAACUUACGAGCUCACAGGGCCAUAUUUGUUAACCGC ---......(.((((((....)))).)))...........((((((((((((......(((...(.((((........)))).)..)))..(.(((.....))))...)))))))))))) ( -28.40) >consensus ___CUUUCCUAACAUCACCUUUCGGUGCCAUAAAAGUUUUGCGGUUAACAAAAUUGAAACGGCGCACGACGCCCACAAAUCGCGAGCGUACGAGCCCGCAGGGCCAUAUUUGUUAACCGC ...............((((....)))).............((((((((((((........((((.....))))......(((........)))((((...))))....)))))))))))) (-30.40 = -29.76 + -0.64)

| Location | 1,560,042 – 1,560,159 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.82 |
| Mean single sequence MFE | -47.58 |
| Consensus MFE | -41.68 |
| Energy contribution | -42.64 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.21 |
| SVM RNA-class probability | 0.999836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1560042 117 - 20766785 GCGGUUAACAAAUGUGGCCCUGCGGGCUCGUACGCUAGCGAUUUGUGGGCGUCGUGCGCCGUUUCAAUUUUGUUAACCGCAAAACUUUUAUGGCACCGAAAGGUGAUGUUAGGAAAG--- ((((((((((((..(((....((((((.((.(((((.((.....)).))))))).)).)))).)))..))))))))))))....(((((....((((....)))).......)))))--- ( -48.70) >DroSec_CAF1 9782 117 - 1 GCGGUUAACAAAUAUGGCCCUGCGGGCUCGCACGCUCGCGAUUUGUGGGCGUCGUGCGCCGUUUCAAUUUUGUUAACCGCAAAACUUUUAUGGCGCCGAAAGGCGAUGUUAGAAAAG--- ((((((((((((..(((....((((((.((.((((((((.....)))))))))).)).)))).)))..)))))))))))).....(((((...((((....))))....)))))...--- ( -54.10) >DroSim_CAF1 11279 120 - 1 GCGGUUAACAAAUAUGGCCCUGCGGGCUCGUACGCUCGCGAUUUGUGGGCGUCGUGCGCCGUUUCAAUUUUGUUAACCGCAAAACUUUUAUGGCACCGAAAGGCGAUGUUAGGAAGGGAU ((((((((((((..(((....((((((.((.((((((((.....)))))))))).)).)))).)))..))))))))))))....(((((.(((((((....))...))))).)))))... ( -52.80) >DroEre_CAF1 10178 116 - 1 GCGGCUAACAAAUAUGGCCCUGCGGGCUCGUAGGUUUGCGAUUUGUGGGCGUCGUGCGCCGUUUCCAUUUUGUUCACCGCAUAACUUUUAUAGCACCGAAAGGUGAGGCCC-AAAGG--- ((((..((((((.((((....((((((.((.(.(((..(.....)..))).))).)).))))..))))))))))..))))....((((.....((((....))))......-)))).--- ( -42.80) >DroYak_CAF1 16342 117 - 1 GCGGUUAACAAAUAUGGCCCUGUGAGCUCGUAAGUUCGCGAUUUGUGGGCAUCGUGCGCCUUUGUAAUUUUGUUAACCGCAAAACUUUUAUGGCACUUAAAGGUAAGGCUCGGAAAG--- ((((((((((((....((((((((((((....))))))))......))))....((((....))))..))))))))))))....(((((..(((.((((....)))))))..)))))--- ( -39.50) >consensus GCGGUUAACAAAUAUGGCCCUGCGGGCUCGUACGCUCGCGAUUUGUGGGCGUCGUGCGCCGUUUCAAUUUUGUUAACCGCAAAACUUUUAUGGCACCGAAAGGUGAUGUUAGGAAAG___ ((((((((((((..(((....((((((.((.((((((((.....)))))))))).)).)))).)))..))))))))))))....(((((.(((((((....)))...)))).)))))... (-41.68 = -42.64 + 0.96)


| Location | 1,560,079 – 1,560,199 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -33.72 |
| Energy contribution | -33.68 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1560079 120 + 20766785 GCGGUUAACAAAAUUGAAACGGCGCACGACGCCCACAAAUCGCUAGCGUACGAGCCCGCAGGGCCACAUUUGUUAACCGCCUGCACUUGACUAUCAUACGUUUAUGAUGUUUGAGCACAA ((((((((((((........((((.....))))......(((........)))((((...))))....)))))))))))).((..((..(.(((((((....))))))).)..))..)). ( -37.20) >DroSec_CAF1 9819 120 + 1 GCGGUUAACAAAAUUGAAACGGCGCACGACGCCCACAAAUCGCGAGCGUGCGAGCCCGCAGGGCCAUAUUUGUUAACCGCUUGCACUUGACUAUCAUACGUUUAUGAUGUUUGGGCACAA ((((((((((((..((......((((((.(((.........)))..)))))).((((...))))))..)))))))))))).(((.((.(((.((((((....))))))))).)))))... ( -45.70) >DroSim_CAF1 11319 120 + 1 GCGGUUAACAAAAUUGAAACGGCGCACGACGCCCACAAAUCGCGAGCGUACGAGCCCGCAGGGCCAUAUUUGUUAACCGCCUGCACUUGACUAUCAUACGUUUAUGAUGUUUGGGCACAA ((((((((((((..((....((((.....))))........(((.((......)).))).....))..)))))))))))).(((.((.(((.((((((....))))))))).)))))... ( -40.80) >DroEre_CAF1 10214 120 + 1 GCGGUGAACAAAAUGGAAACGGCGCACGACGCCCACAAAUCGCAAACCUACGAGCCCGCAGGGCCAUAUUUGUUAGCCGCCUGCACUUGACUAUCAUGCGUUUAUGAUGUUUGGGCUCUU (((((.........(....)((((.....)))).....)))))........(((((((((((((...........))..)))))....(((.((((((....))))))))).)))))).. ( -38.40) >DroYak_CAF1 16379 120 + 1 GCGGUUAACAAAAUUACAAAGGCGCACGAUGCCCACAAAUCGCGAACUUACGAGCUCACAGGGCCAUAUUUGUUAACCGCUCGCACUUGUCUACCAUACGGUUAUGAUGUUUCUGCUCGU ((((((((((((......(((...(.((((........)))).)..)))..(.(((.....))))...))))))))))))..(((..((((.(((....)))...))))....))).... ( -32.50) >consensus GCGGUUAACAAAAUUGAAACGGCGCACGACGCCCACAAAUCGCGAGCGUACGAGCCCGCAGGGCCAUAUUUGUUAACCGCCUGCACUUGACUAUCAUACGUUUAUGAUGUUUGGGCACAA ((((((((((((........((((.....))))......(((........)))((((...))))....))))))))))))..((....(((.((((((....)))))))))...)).... (-33.72 = -33.68 + -0.04)

| Location | 1,560,079 – 1,560,199 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -43.98 |
| Consensus MFE | -36.02 |
| Energy contribution | -37.30 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1560079 120 - 20766785 UUGUGCUCAAACAUCAUAAACGUAUGAUAGUCAAGUGCAGGCGGUUAACAAAUGUGGCCCUGCGGGCUCGUACGCUAGCGAUUUGUGGGCGUCGUGCGCCGUUUCAAUUUUGUUAACCGC (((..((...((((((((....)))))).))..))..)))((((((((((((..(((....((((((.((.(((((.((.....)).))))))).)).)))).)))..)))))))))))) ( -46.20) >DroSec_CAF1 9819 120 - 1 UUGUGCCCAAACAUCAUAAACGUAUGAUAGUCAAGUGCAAGCGGUUAACAAAUAUGGCCCUGCGGGCUCGCACGCUCGCGAUUUGUGGGCGUCGUGCGCCGUUUCAAUUUUGUUAACCGC (((..(....((((((((....)))))).))...)..)))((((((((((((..(((....((((((.((.((((((((.....)))))))))).)).)))).)))..)))))))))))) ( -49.00) >DroSim_CAF1 11319 120 - 1 UUGUGCCCAAACAUCAUAAACGUAUGAUAGUCAAGUGCAGGCGGUUAACAAAUAUGGCCCUGCGGGCUCGUACGCUCGCGAUUUGUGGGCGUCGUGCGCCGUUUCAAUUUUGUUAACCGC (((..(....((((((((....)))))).))...)..)))((((((((((((..(((....((((((.((.((((((((.....)))))))))).)).)))).)))..)))))))))))) ( -48.50) >DroEre_CAF1 10214 120 - 1 AAGAGCCCAAACAUCAUAAACGCAUGAUAGUCAAGUGCAGGCGGCUAACAAAUAUGGCCCUGCGGGCUCGUAGGUUUGCGAUUUGUGGGCGUCGUGCGCCGUUUCCAUUUUGUUCACCGC ..((((((....(((((......)))))........(((((..((((.......)))))))))))))))...(((..((((..((..((((.(....).))))..))..))))..))).. ( -41.10) >DroYak_CAF1 16379 120 - 1 ACGAGCAGAAACAUCAUAACCGUAUGGUAGACAAGUGCGAGCGGUUAACAAAUAUGGCCCUGUGAGCUCGUAAGUUCGCGAUUUGUGGGCAUCGUGCGCCUUUGUAAUUUUGUUAACCGC ....(((.....((((((....)))))).......)))..((((((((((((....((((((((((((....))))))))......))))....((((....))))..)))))))))))) ( -35.10) >consensus UUGUGCCCAAACAUCAUAAACGUAUGAUAGUCAAGUGCAGGCGGUUAACAAAUAUGGCCCUGCGGGCUCGUACGCUCGCGAUUUGUGGGCGUCGUGCGCCGUUUCAAUUUUGUUAACCGC .((..(......((((((....))))))......)..)).((((((((((((..(((....((((((.((.((((((((.....)))))))))).)).)))).)))..)))))))))))) (-36.02 = -37.30 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:53 2006