
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,172,219 – 8,172,370 |
| Length | 151 |
| Max. P | 0.954042 |

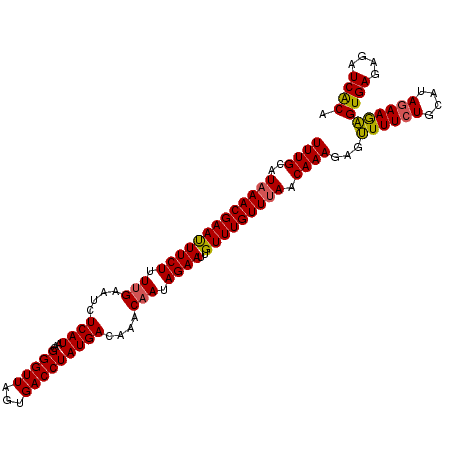
| Location | 8,172,219 – 8,172,330 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -21.14 |
| Energy contribution | -20.78 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
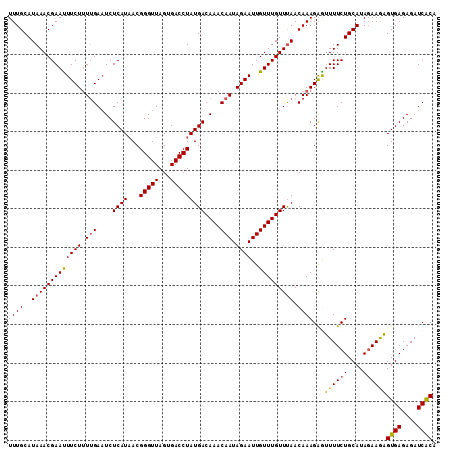
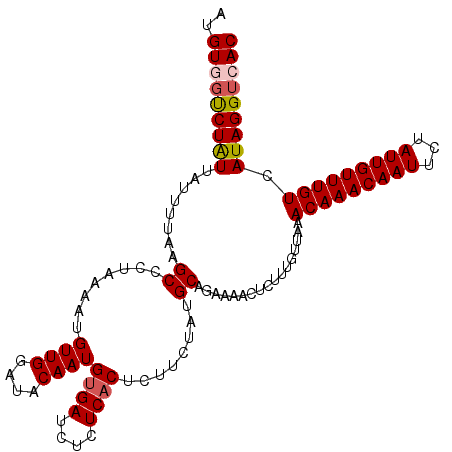
>2R_DroMel_CAF1 8172219 111 + 20766785 UUUGCAUAAACGAAUUUCUAUUGAAUCUCAUAACGGGUUAGUGACCUAUGACAAACAAUAGAAUUGUUUGUUUAACAAAGAAUUUUCUGCAUAGAAGAGUGAGAGAUCACA ((((..(((((((((((((((((....((((...(((((...)))))))))....))))))))..))))))))).))))...((((((....))))))((((....)))). ( -26.30) >DroSec_CAF1 3094 111 + 1 UUUGCAUAAACGAAUUUCUUUUGAAUCUCAUAACGGGUUAGUGACCUAUGACAAACAAUAGAAUUGUUUGUUUAACAAAGAGUUUUCUGCAUACAAGAGUGAGAGAUCACA ..((((.((((((..(((....)))..)).....(((((...)))))..((((((((((...)))))))))).........))))..)))).......((((....)))). ( -22.80) >DroSim_CAF1 3121 111 + 1 UUUGCAUAAACGAAUUUCUUUUGAAUCUCAUAACGGGUUAGUGACCUAUGACAAACAAUAGAAUUGUUUGUUUAACAAAGAAUUUUCUGCAUAGAAGAGUGAGAGAUCACA ((((..(((((((((((((.(((....((((...(((((...)))))))))....))).))))..))))))))).))))...((((((....))))))((((....)))). ( -22.40) >DroEre_CAF1 3113 111 + 1 UUUGCAUAAACGAACUUCUUUUGAUUCUCAUUACGGGUUAGUGACCUAUGACAAACAAUAGAAUUGUUUGUUUAACAAAGAGUUUUCUGCAUAGAAAAGUGAGAGAUCGCA ..(((.(((((((((((((.(((.((.((((...(((((...))))))))).)).))).))))..))))))))).......(.(((((.(((......)))))))).)))) ( -24.60) >DroYak_CAF1 3139 111 + 1 UUUGCAUAAACGAACUUCUUUGGAAUCUCAUAACGGGUUAGUGACCUAUGACAAACAAUAGAAUUGUUUGUUCAACAAAGGGAUUUCUGCAUAGAAAUGUGAGAGAUCACA ................((((((............(((((...))))).(((((((((((...)))))))).))).))))))(((((((.((((....)))))))))))... ( -28.10) >consensus UUUGCAUAAACGAAUUUCUUUUGAAUCUCAUAACGGGUUAGUGACCUAUGACAAACAAUAGAAUUGUUUGUUUAACAAAGAGUUUUCUGCAUAGAAGAGUGAGAGAUCACA ((((..(((((((((((((.(((....((((...(((((...)))))))))....))).))))..))))))))).))))...((((((....))))))((((....)))). (-21.14 = -20.78 + -0.36)

| Location | 8,172,219 – 8,172,330 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
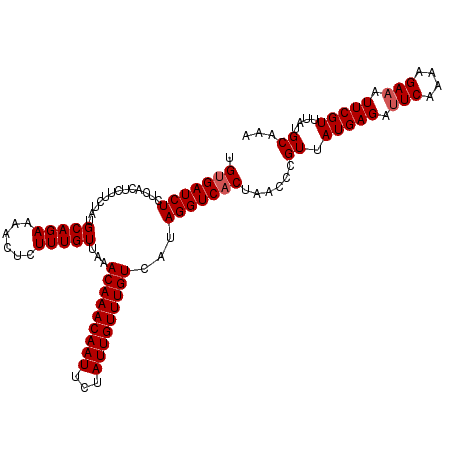
| Reading direction | reverse |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8172219 111 - 20766785 UGUGAUCUCUCACUCUUCUAUGCAGAAAAUUCUUUGUUAAACAAACAAUUCUAUUGUUUGUCAUAGGUCACUAACCCGUUAUGAGAUUCAAUAGAAAUUCGUUUAUGCAAA .((((....)))).......((((.(((.....(((((.....)))))((((((((..(.((((((((.....)))...))))).)..)))))))).....))).)))).. ( -23.20) >DroSec_CAF1 3094 111 - 1 UGUGAUCUCUCACUCUUGUAUGCAGAAAACUCUUUGUUAAACAAACAAUUCUAUUGUUUGUCAUAGGUCACUAACCCGUUAUGAGAUUCAAAAGAAAUUCGUUUAUGCAAA .((((....)))).......((((..((((...(((((.....)))))((((.(((..(.((((((((.....)))...))))).)..))).))))....)))).)))).. ( -21.50) >DroSim_CAF1 3121 111 - 1 UGUGAUCUCUCACUCUUCUAUGCAGAAAAUUCUUUGUUAAACAAACAAUUCUAUUGUUUGUCAUAGGUCACUAACCCGUUAUGAGAUUCAAAAGAAAUUCGUUUAUGCAAA .(((((((.............(((((......)))))...(((((((((...)))))))))...)))))))......((.(((((.(((....))).)))))....))... ( -20.10) >DroEre_CAF1 3113 111 - 1 UGCGAUCUCUCACUUUUCUAUGCAGAAAACUCUUUGUUAAACAAACAAUUCUAUUGUUUGUCAUAGGUCACUAACCCGUAAUGAGAAUCAAAAGAAGUUCGUUUAUGCAAA ((((((((.....((((((....))))))...........(((((((((...)))))))))...)))))..........((((((..((....))..))))))...))).. ( -18.90) >DroYak_CAF1 3139 111 - 1 UGUGAUCUCUCACAUUUCUAUGCAGAAAUCCCUUUGUUGAACAAACAAUUCUAUUGUUUGUCAUAGGUCACUAACCCGUUAUGAGAUUCCAAAGAAGUUCGUUUAUGCAAA (((((....)))))......((((.((((..((((.(((..((((((((...))))))))((((((((.....)))...))))).....))).))))...)))).)))).. ( -21.60) >consensus UGUGAUCUCUCACUCUUCUAUGCAGAAAACUCUUUGUUAAACAAACAAUUCUAUUGUUUGUCAUAGGUCACUAACCCGUUAUGAGAUUCAAAAGAAAUUCGUUUAUGCAAA .(((((((.............(((((......)))))...(((((((((...)))))))))...)))))))......((.(((((.(((....))).)))))....))... (-18.28 = -18.68 + 0.40)


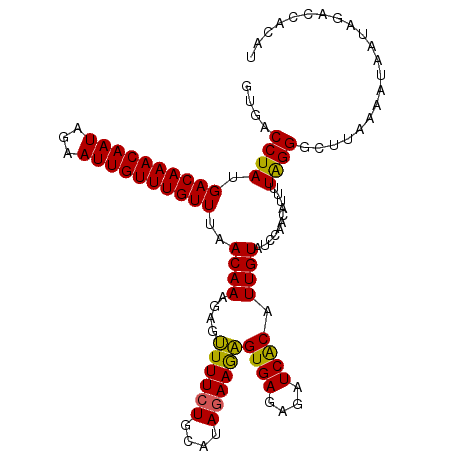
| Location | 8,172,259 – 8,172,370 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -19.36 |
| Energy contribution | -18.68 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8172259 111 + 20766785 GUGACCUAUGACAAACAAUAGAAUUGUUUGUUUAACAAAGAAUUUUCUGCAUAGAAGAGUGAGAGAUCACAUUGUAUCCAACAUUUUAGGGCUUAAAAUAAUAGACAACAU ....((((.((((((((((...))))))))))..((((....((((((....))))))((((....)))).))))...........))))..................... ( -19.80) >DroSec_CAF1 3134 111 + 1 GUGACCUAUGACAAACAAUAGAAUUGUUUGUUUAACAAAGAGUUUUCUGCAUACAAGAGUGAGAGAUCACAUUGUAUUCAACAUUUUAGGGCUUAUUAUAAUAGGCCACAU (((.((((.((((((((((...))))))))))...............((.((((((..((((....)))).)))))).))......))))((((((....))))))))).. ( -27.70) >DroSim_CAF1 3161 111 + 1 GUGACCUAUGACAAACAAUAGAAUUGUUUGUUUAACAAAGAAUUUUCUGCAUAGAAGAGUGAGAGAUCACAUUGUAUCCAACAUUUUAGGGCUUAAAAUAAUAGGCGACAU ....((((.((((((((((...))))))))))..((((....((((((....))))))((((....)))).))))...........))))(((((......)))))..... ( -23.80) >DroEre_CAF1 3153 111 + 1 GUGACCUAUGACAAACAAUAGAAUUGUUUGUUUAACAAAGAGUUUUCUGCAUAGAAAAGUGAGAGAUCGCAUUGUAUCCAACAUUUUAGGGCUUAAAAUAACAGACCACAU (((..((..((((((((((...)))))))))).........(((((..((.(((((..((((....))))..(((.....))))))))..))..)))))...))..))).. ( -19.60) >DroYak_CAF1 3179 111 + 1 GUGACCUAUGACAAACAAUAGAAUUGUUUGUUCAACAAAGGGAUUUCUGCAUAGAAAUGUGAGAGAUCACAUUGUGGCCAACAUUUUGGGGCUUCAAAUAACAGAACACAU (((.(((.(((((((((((...)))))))))....)).)))(((((((.((((....)))))))))))...(((.((((..........)))).))).........))).. ( -30.70) >consensus GUGACCUAUGACAAACAAUAGAAUUGUUUGUUUAACAAAGAGUUUUCUGCAUAGAAGAGUGAGAGAUCACAUUGUAUCCAACAUUUUAGGGCUUAAAAUAAUAGACCACAU ....((((.((((((((((...))))))))))..((((....((((((....))))))((((....)))).))))...........))))..................... (-19.36 = -18.68 + -0.68)


| Location | 8,172,259 – 8,172,370 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.34 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.672497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8172259 111 - 20766785 AUGUUGUCUAUUAUUUUAAGCCCUAAAAUGUUGGAUACAAUGUGAUCUCUCACUCUUCUAUGCAGAAAAUUCUUUGUUAAACAAACAAUUCUAUUGUUUGUCAUAGGUCAC .(((.(((((.(((((((.....))))))).))))))))..(((((((.............(((((......)))))...(((((((((...)))))))))...))))))) ( -22.70) >DroSec_CAF1 3134 111 - 1 AUGUGGCCUAUUAUAAUAAGCCCUAAAAUGUUGAAUACAAUGUGAUCUCUCACUCUUGUAUGCAGAAAACUCUUUGUUAAACAAACAAUUCUAUUGUUUGUCAUAGGUCAC ..(((((((((..(((((((........(((...((((((.((((....))))..)))))))))........))))))).(((((((((...))))))))).))))))))) ( -27.79) >DroSim_CAF1 3161 111 - 1 AUGUCGCCUAUUAUUUUAAGCCCUAAAAUGUUGGAUACAAUGUGAUCUCUCACUCUUCUAUGCAGAAAAUUCUUUGUUAAACAAACAAUUCUAUUGUUUGUCAUAGGUCAC .(((..((...(((((((.....)))))))..))..)))..(((((((.............(((((......)))))...(((((((((...)))))))))...))))))) ( -20.60) >DroEre_CAF1 3153 111 - 1 AUGUGGUCUGUUAUUUUAAGCCCUAAAAUGUUGGAUACAAUGCGAUCUCUCACUUUUCUAUGCAGAAAACUCUUUGUUAAACAAACAAUUCUAUUGUUUGUCAUAGGUCAC .((((.(((..(((((((.....)))))))..)))))))....(((((.....((((((....))))))...........(((((((((...)))))))))...))))).. ( -19.80) >DroYak_CAF1 3179 111 - 1 AUGUGUUCUGUUAUUUGAAGCCCCAAAAUGUUGGCCACAAUGUGAUCUCUCACAUUUCUAUGCAGAAAUCCCUUUGUUGAACAAACAAUUCUAUUGUUUGUCAUAGGUCAC ..(((((((((.....((((((..........)))....((((((....)))))))))...))))))...(((....(((.((((((((...))))))))))).))).))) ( -23.90) >consensus AUGUGGUCUAUUAUUUUAAGCCCUAAAAUGUUGGAUACAAUGUGAUCUCUCACUCUUCUAUGCAGAAAACUCUUUGUUAAACAAACAAUUCUAUUGUUUGUCAUAGGUCAC ..(((((((((........((........((((....))))((((....))))........)).................(((((((((...))))))))).))))))))) (-17.02 = -17.34 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:10 2006