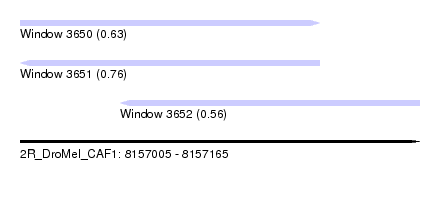
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,157,005 – 8,157,165 |
| Length | 160 |
| Max. P | 0.762130 |

| Location | 8,157,005 – 8,157,125 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -19.11 |
| Energy contribution | -20.25 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8157005 120 + 20766785 AACACGCCCAGGAAGAUGUUCUUGACCAGCUUCCACUUGCACACGCCCAGGCGGUCGGGCCACAGAUAGACGGUCUCCACGAAACUGGGCACAAAGAUGCCCAGCAGCGAGAAGAACACG .....(((((((((....)))))((((.((........))....((....))))))))))..........((.(((.(.((...(((((((......)))))))...)).).)))...)) ( -38.20) >DroVir_CAF1 7487 120 + 1 AAAACGCCCAAGAGAAUGUUCUUGAUCAGCUUCCACUUGCAGCAGCCCAGACGAUCGGGCCACAGAUAUACAGUCUCCACAAAGCUGGGCACAAAGAUGCCCAGCAGCGAGAAGAAGACG ........((((((....))))))....(((((..(((((....((((........))))...((((.....)))).......((((((((......)))))))).)))))..)))).). ( -38.80) >DroPse_CAF1 4126 120 + 1 AGAAUGCCCAAAAUAAUAUUCUUUACCAUCUUCCAAUUGCACCAACCCAGACGAUCGGGAUAAAGGUAAACGGUCUCAACGAAGCUGGGAACAAAAAUGCCUAGCAGUGAGAAGAACACG ((((((..........))))))......(((((..(((((.....(((........)))....(((((.....(((((.......))))).......))))).)))))..)))))..... ( -25.20) >DroGri_CAF1 5016 120 + 1 AAGAUGCCCAAAAGAACGUUCUUGAUCAGCUUCCAUUUGCACCAGCCGAGGCGAUCAGGCCACAGAUACACAGUCUCAAUGAAGCUGGGCACAAAUAUGCCCAGCAGCGAGAAAAACACA ....(((.........(((((((((((.(((((.....((....)).))))))))))))....((((.....)))).))))..((((((((......)))))))).)))........... ( -33.00) >DroWil_CAF1 5913 117 + 1 AAUACGGCAAGCAAAAUAUUCUUAAUCAGUUUCCAUUUGCACCAUCCCAA---AUUGGGCCACAAGUAGACAGUCUCCACAACACUGGGCACAAAGAUACCCAGCAAGGAGAAGAACACA .....(((..(((((.....((.....))......))))).(((......---..))))))....((......(((((......(((((..........)))))...))))).....)). ( -23.60) >DroMoj_CAF1 7338 120 + 1 AAAACUCCCAACAAUAUGUUCUUGAUAAGCUUCCAUUUGCACCAGCCCAGACGAUCGGGCCACAGGUACACGGUCUCCACGAAGCUGGGCACAAAGAUGCCCAGCAGCGAAAAGAAGACG ................(((.((((....((........))....((((........))))..)))).)))..(((((..((..((((((((......))))))))..))....).)))). ( -34.30) >consensus AAAACGCCCAAAAAAAUGUUCUUGAUCAGCUUCCAUUUGCACCAGCCCAGACGAUCGGGCCACAGAUACACAGUCUCCACGAAGCUGGGCACAAAGAUGCCCAGCAGCGAGAAGAACACG ...................................(((((....((((........)))).......................((((((((......)))))))).)))))......... (-19.11 = -20.25 + 1.14)

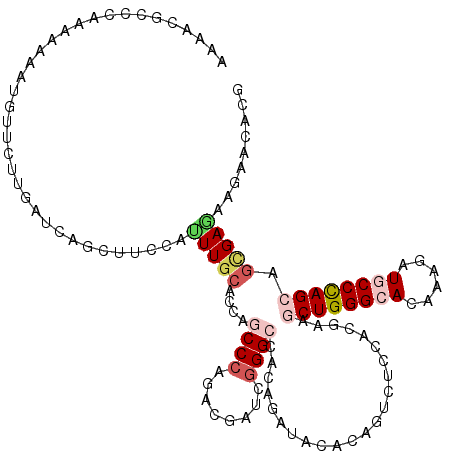
| Location | 8,157,005 – 8,157,125 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -27.27 |
| Energy contribution | -27.42 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.762130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8157005 120 - 20766785 CGUGUUCUUCUCGCUGCUGGGCAUCUUUGUGCCCAGUUUCGUGGAGACCGUCUAUCUGUGGCCCGACCGCCUGGGCGUGUGCAAGUGGAAGCUGGUCAAGAACAUCUUCCUGGGCGUGUU ((.((.((((.((..(((((((((....)))))))))..)).))))))))..........((((((((((....))).))......(((((.((........)).))))))))))..... ( -46.40) >DroVir_CAF1 7487 120 - 1 CGUCUUCUUCUCGCUGCUGGGCAUCUUUGUGCCCAGCUUUGUGGAGACUGUAUAUCUGUGGCCCGAUCGUCUGGGCUGCUGCAAGUGGAAGCUGAUCAAGAACAUUCUCUUGGGCGUUUU ((((..((((.(((((((((((((....)))))))))..((((....).........(..(((((......)))))..).))))))))))).....(((((......))))))))).... ( -45.60) >DroPse_CAF1 4126 120 - 1 CGUGUUCUUCUCACUGCUAGGCAUUUUUGUUCCCAGCUUCGUUGAGACCGUUUACCUUUAUCCCGAUCGUCUGGGUUGGUGCAAUUGGAAGAUGGUAAAGAAUAUUAUUUUGGGCAUUCU .(((((((((((((.(((.((.((....)).)).)))...).))))(((((((.((.....(((((((.....)))))).).....)).))))))).))))))))............... ( -28.30) >DroGri_CAF1 5016 120 - 1 UGUGUUUUUCUCGCUGCUGGGCAUAUUUGUGCCCAGCUUCAUUGAGACUGUGUAUCUGUGGCCUGAUCGCCUCGGCUGGUGCAAAUGGAAGCUGAUCAAGAACGUUCUUUUGGGCAUCUU ...((((((.((((((((((((((....)))))))))((((((.....((..(..(((.(((......))).)))...)..))))))))))).))..))))))((((....))))..... ( -38.70) >DroWil_CAF1 5913 117 - 1 UGUGUUCUUCUCCUUGCUGGGUAUCUUUGUGCCCAGUGUUGUGGAGACUGUCUACUUGUGGCCCAAU---UUGGGAUGGUGCAAAUGGAAACUGAUUAAGAAUAUUUUGCUUGCCGUAUU .((((((((((((.((((((((((....))))))))))....)))))...((((.(((((.(((...---..)))....))))).)))).........)))))))............... ( -33.80) >DroMoj_CAF1 7338 120 - 1 CGUCUUCUUUUCGCUGCUGGGCAUCUUUGUGCCCAGCUUCGUGGAGACCGUGUACCUGUGGCCCGAUCGUCUGGGCUGGUGCAAAUGGAAGCUUAUCAAGAACAUAUUGUUGGGAGUUUU .((((((....((..(((((((((....)))))))))..)).))))))..((((((...((((((......))))))))))))....(((((((.((((.((....)).))))))))))) ( -48.40) >consensus CGUGUUCUUCUCGCUGCUGGGCAUCUUUGUGCCCAGCUUCGUGGAGACCGUCUACCUGUGGCCCGAUCGUCUGGGCUGGUGCAAAUGGAAGCUGAUCAAGAACAUUUUCUUGGGCGUUUU .(((((((((((((.(((((((((....)))))))))...)).))))((((.((((...((((((......))))))))))...))))..........)))))))............... (-27.27 = -27.42 + 0.15)
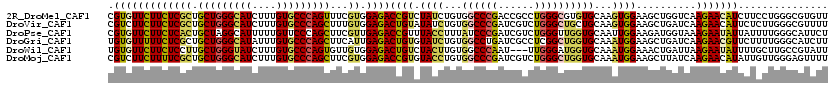
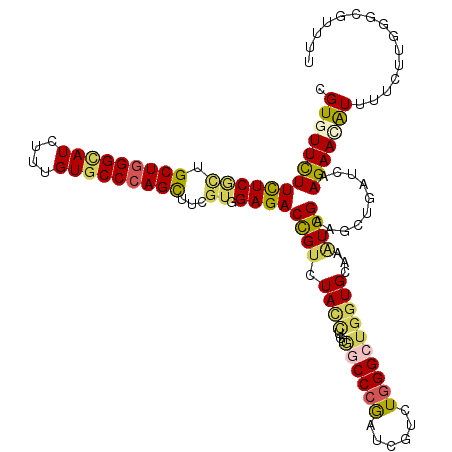
| Location | 8,157,045 – 8,157,165 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.94 |
| Mean single sequence MFE | -43.22 |
| Consensus MFE | -28.93 |
| Energy contribution | -29.60 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8157045 120 - 20766785 CUAUCCCCAACCUGGAACCCUUCAUCAGUCUGGUGGGUGCCGUGUUCUUCUCGCUGCUGGGCAUCUUUGUGCCCAGUUUCGUGGAGACCGUCUAUCUGUGGCCCGACCGCCUGGGCGUGU ........(((.(((.((((.(((......))).)))).))).)))((((.((..(((((((((....)))))))))..)).))))((((((((...((((.....)))).)))))).)) ( -44.40) >DroVir_CAF1 7527 120 - 1 GCAUUCCCAACCUGGAACCGUUCAUCAGUUUGGUGGGCGCCGUCUUCUUCUCGCUGCUGGGCAUCUUUGUGCCCAGCUUUGUGGAGACUGUAUAUCUGUGGCCCGAUCGUCUGGGCUGCU ((.((((......))))..(((((((.....))))))))).((((((....((..(((((((((....)))))))))..)).)))))).........(..(((((......)))))..). ( -48.80) >DroPse_CAF1 4166 120 - 1 CUAUUCCCAACCUGGAACCGUUCAUCAGCUUGGUGGGCGCCGUGUUCUUCUCACUGCUAGGCAUUUUUGUUCCCAGCUUCGUUGAGACCGUUUACCUUUAUCCCGAUCGUCUGGGUUGGU ......((((((..((..((((((((.....))))))))..(((..(.((((((.(((.((.((....)).)).)))...).)))))..)..)))..............))..)))))). ( -33.90) >DroGri_CAF1 5056 120 - 1 GCAUACCCAAUCUGGAGCCGUUCAUCAGUUUGGUGGGCGCUGUGUUUUUCUCGCUGCUGGGCAUAUUUGUGCCCAGCUUCAUUGAGACUGUGUAUCUGUGGCCUGAUCGCCUCGGCUGGU (((((.((.....))(((.(((((((.....)))))))))).......(((((..(((((((((....))))))))).....))))).)))))..(((.(((......))).)))..... ( -40.90) >DroWil_CAF1 5953 117 - 1 CCAUUCCCGACUUGGAACCGUUCAUCAGUUUGGUGGGAGCUGUGUUCUUCUCCUUGCUGGGUAUCUUUGUGCCCAGUGUUGUGGAGACUGUCUACUUGUGGCCCAAU---UUGGGAUGGU ((((.(((((.((((..(((..((.((((((.....))))))))..).(((((.((((((((((....))))))))))....)))))............)).)))).---))))))))). ( -43.60) >DroMoj_CAF1 7378 120 - 1 GCAUUCCCAAUCUGGAGCCAUUCAUCAGUCUCGUGGGCGCCGUCUUCUUUUCGCUGCUGGGCAUCUUUGUGCCCAGCUUCGUGGAGACCGUGUACCUGUGGCCCGAUCGUCUGGGCUGGU ....(((......)))((((...........((..(((((.((((((....((..(((((((((....)))))))))..)).)))))).)))..))..))(((((......))))))))) ( -47.70) >consensus CCAUUCCCAACCUGGAACCGUUCAUCAGUUUGGUGGGCGCCGUGUUCUUCUCGCUGCUGGGCAUCUUUGUGCCCAGCUUCGUGGAGACCGUCUACCUGUGGCCCGAUCGUCUGGGCUGGU .....((((..((((.........)))).....))))........(((((.((..(((((((((....)))))))))..)).)))))............((((((......))))))... (-28.93 = -29.60 + 0.67)

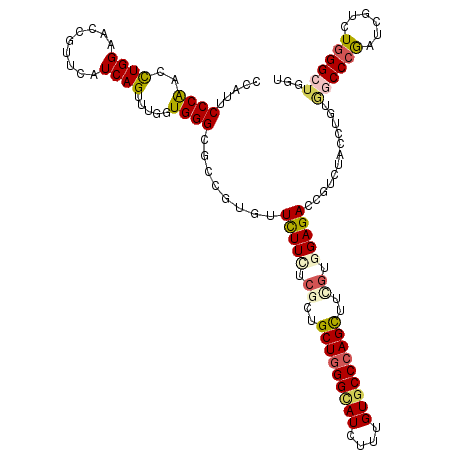
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:04 2006